
miR-31: the double-edged sword of CD8 T lymphocytes
MicroRNAs (miRNAs) are small regulatory molecules that control gene expression. During the immune response, both initiation and resolution of the response must be tightly regulated to achieve a successful clearance of the infectious agent, avoiding excessive activation that could be detrimental for the host. T cell activation comprises a plethora of signaling pathways that promote expansion and differentiation of the T cell repertoire into effector and memory cells. Moreover, T cells can become exhausted and even dysfunctional under certain circumstances. However, the precise mechanisms that control these processes are only starting to be elucidated. Not surprisingly, microRNAs have important roles in the control of T cell biology and function.
The ablation of the miRNA-producing enzyme Dicer can impact both the CD8 T cell development and effector function (1-3). Moreover, several miRNAs have been described to be regulated in CD8 T cells playing important roles in their biology (4,5). Additionally, upon T cell activation, global miRNA regulation of gene expression changes dramatically due to different processes like argonaute proteins degradation (6), shortening of mRNA targets 3’UTR (7) and even post-transcriptional non templated uridylation of miRNAs (8). In the recently published work in Nature Immunology by Moffett and colleagues (9), miR-31 function in CD8 T cells is uncovered for the first time. Authors describe that the expression of miR-31 is highly increased during CD8 T cell activation being controlled by nuclear factor of activated T cells (NFAT). Moreover, they show the implication of this miRNA in limiting CD8 T cell function that ultimately causes the inability of the host to resolve the chronic phase of lymphocytic choriomeningitis virus (LCMV) clone 13 infection (9).
After already a quarter of a century of miRNA research, mRNA target prediction and validation are still very challenging subjects. The difficulties are originated mainly by the incomplete matching of sequences between miRNA seed sequence and the target mRNA 3’UTR (10). In their study, Moffett et al. use two approaches to experimentally predict the miR-31 mRNA targets (9). First they identify its direct targets by comparing mRNA expression of control and miR-31 overexpressing cells by microarrays and subsequent validation by 3’UTR luciferase reporter assays. Secondly, they use miR-31 deficient mice to assess the gene expression changes in the absence of this miRNA by RNA sequencing and validation by qPCR and flow cytometry. Interestingly, in silico study by ingenuity pathway analysis of the targets identified in the first approach, leads them to type I interferon (IFN) pathway. Accordingly, they then study the T cell function of miR-31 deficient cells finding that T cell dysfunction is decreased and T cell effector molecules increased in the miR-31 deficient cells in an IFN-β treatment dependent fashion. The main target of miR-31 responsible for this effect is the phosphatase Ppp6c that would have been inhibiting type I IFN signaling otherwise. The consequences of this signaling modulation is linked to the differential expression of genes related to T cell dysfunction that impact on the CD8 T cell capacity to deal with infection (Figure 1).
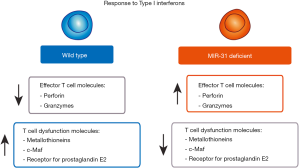
Although miRNA levels can change very rapidly upon T cell activation (9,11,12), the actual impact of these changes in the levels of the proteins regulated by miRNAs is often delayed in time. Target proteins half lives as well as transcription rates of both mRNA and miRNA are variable parameters that influence the time when the outcome of miRNA levels changes will be evident. The study by Moffett et al. represents a nice in vivo example of the delay of miRNA effect. Since miR-31 is induced very rapidly after T cell activation, it would be expectable that its deficiency would affect the acute infection of influenza virus or LCMV. However, the difference between wild type and deficient mice starts to be evident only at the chronic phase of LCMV infection (9) (Figure 2). These findings also confirm that miRNA-driven regulation comprises very intricate spatiotemporal regulatory networks.
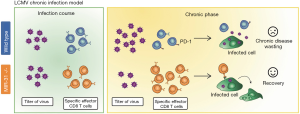
During viral chronic infection, the absence of miR-31 helps CD8 T cells to mount a more efficient response and prevents them to be exhausted as wild types. This poses an intriguing question that remains unsolved: why would the host upregulate this miRNA during infection if it is not beneficial but detrimental in this context? The most plausible explanation is that miR-31 probably represents a mechanism to prevent excessive T cell response in other situations which in this particular case has turned against the host.
Collectively, the findings by Moffett and coworkers uncovered a relevant mechanism of control of T cell dysfunction that could be interpreted beyond the context of infection. T cell dysfunction and exhaustion is a common feature of cancer that is often promoted by the tumor itself. The identification of miR-31 as a target to prevent this CD8 T cell aberrant behavior opens important new avenues that would be worth it to explore in order to develop new therapeutic approaches against tumors. Conversely, promoting a less efficient immune response is the goal of autoimmunity treatment that could be potentially be accomplished by boosting miR-31 expression specially in diseases dependent on type I IFN signaling like systemic lupus erythematosus or multiple sclerosis (13) among others.
Acknowledgments
Funding: This work was supported by grants to F Sánchez-Madrid (SAF2014-55579-R; ERC-2011-AdG 294340-GENTRIS and CIBER CARDIOVASCULAR). C Gutiérrez-Vázquez is supported by an Alfonso Martín Escudero Foundation postdoctoral fellowship.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Yikun Yao (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.08.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cobb BS, Nesterova TB, Thompson E, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med 2005;201:1367-73. [Crossref] [PubMed]
- Muljo SA, Ansel KM, Kanellopoulou C, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med 2005;202:261-9. [Crossref] [PubMed]
- Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci U S A 2010;107:21629-34. [Crossref] [PubMed]
- Liang Y, Pan HF, Ye DQ. microRNAs function in CD8+T cell biology. J Leukoc Biol 2015;97:487-97. [Crossref] [PubMed]
- Trifari S, Pipkin ME, Bandukwala HS, et al. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci U S A 2013;110:18608-13. [Crossref] [PubMed]
- Bronevetsky Y, Villarino AV, Eisley CJ, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med 2013;210:417-32. [Crossref] [PubMed]
- Sandberg R, Neilson JR, Sarma A, et al. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science 2008;320:1643-7. [Crossref] [PubMed]
- Gutiérrez-Vázquez C, Enright AJ, Rodríguez-Galán A, et al. 3' Uridylation controls mature microRNA turnover during CD4 T-cell activation. RNA 2017;23:882-91. [Crossref] [PubMed]
- Moffett HF, Cartwright ANR, Kim HJ, et al. The microRNA miR-31 inhibits CD8+ T cell function in chronic viral infection. Nat Immunol 2017;18:791-9. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4. [PubMed]
- Gutiérrez-Vázquez C, Rodríguez-Galán A, Fernández-Alfara M, et al. miRNA profiling during antigen-dependent T cell activation: A role for miR-132-3p. Sci Rep 2017;7:3508. [Crossref] [PubMed]
- Monticelli S, Ansel KM, Xiao C, et al. MicroRNA profiling of the murine hematopoietic system. genome Biol 2005;6:R71.
- Cepok S, Schreiber H, Hoffmann S, et al. Enhancement of chemokine expression by interferon beta therapy in patients with multiple sclerosis. Arch Neurol 2009;66:1216-23. [Crossref] [PubMed]
Cite this article as: Gutiérrez-Vázquez C, Sánchez-Madrid F. miR-31: the double-edged sword of CD8 T lymphocytes. Non-coding RNA Investig 2017;1:9.


