Non-coding RNAs in exercise
Introduction
The benefits of exercise on human health have been recognized for centuries (1-3). Exercise is an important component in the prevention and treatment of multiple chronic diseases including cardiovascular disease (CVD) (e.g., heart failure), metabolic disorders (e.g., diabetes, obesity) (4), neurological diseases (e.g., Alzheimer’s disease) (5), musculoskeletal disorders (e.g., muscular atrophy) (6) and cancer (7). Exercise challenges whole-body homeostasis and provokes widespread perturbations in numerous tissues and organs following physical training (8). Movement begins via contracting myofibers and triggers a burst of metabolic and morphological adaptations in skeletal muscle. To meet the increased metabolic activity of contracting skeletal muscle, systematically coordinated organs (cardiovascular, respiratory, neural, and endocrine) supply the contracting muscles with more fuel and O2 to sustain a given level of activity (9). Non-coding RNAs (ncRNAs) may act as mediators of physiological processes linked to exercise adaptation. This review aims to summarize these novel molecules and may eventually help develop ncRNAs as therapeutic targets for improving exercise capacity in patients with heart failure and other diseases (Figure 1).
ncRNAs
ncRNAs, a group of small non-protein coding RNAs, account for the majority of all RNA transcripts in the cell. ncRNAs are a diverse group of endogenous RNA-based molecules, which include short (~22 nucleotides) microRNAs (miRNAs, miRs) and long non-coding RNAs (lncRNAs) (of >200 nucleotides) (10).
miRNAs, function as post-transcriptional repressors of gene expression by direct repression or mRNA decay in a sequence-dependent manner. As a class of regulatory molecules, miRNAs are intracellular mediators of adaptive processes, including muscle atrophy (11), CVD (12), aging (13) and cancer (14). The maturation process of miRNAs is an enormous and complex network requiring co-ordination of pri-miRNA transcription (15,16). This process includes cleavage by endonucleases, exportation from nucleus to cytoplasm (17), additional cleavage and then incorporation into the RNA-induced silencing complex (RISC) to inhibit target genes mRNA translation or promote mRNA degradation via canonical base-pairing to the 3’-untranslated (3’UTR) region (18). Most miRNAs are present in the genome as the form of single copy, or multiple copies or cluster (19). miR-1, miR-133a and miR-206 were highly enriched in both heart and skeletal muscle in human and mouse (20,21). Subsequent studies confirmed that a cluster of miRNAs including miR-1, miR-133a, miR-133b, miR-206, miR-208, miR-208b, miR-486, and miR-499 (22) were highly enriched in skeletal muscle, and have therefore been termed “myomiRs” (23). Growing evidences have showed that these muscle-specific miRNAs, along with other miRNAs affect exercise adjustment, suggesting the significance role of miRNAs in response to physical work (24).
Besides long protein-coding mRNAs and short noncoding transcripts, lncRNAs also play important roles in the regulation of cellular processes (25). The majority of lncRNAs are transcribed by RNA polymerase II and function as an additional regulator of the genome via multiple mechanisms (26). As transcriptional regulators, lncRNAs can modulate transcription initiation, elongation and termination (27). In addition, lncRNAs can also mediate gene expression in post-transcriptional processes such as splicing, transport, translation, stability of mRNA, and subcellular localization of proteins (28-30). Indeed, a recent study has demonstrated that lncRNAs could act as cofactors, competitors, and/or decoys of RNA-binding proteins and miRNAs (31). A better understanding of how lncRNAs function can be utilized for locus-specific manipulation of gene expression is an important goal for current research efforts.
miRNAs in exercise
Recently, miRNAs adjustments in skeletal muscle (32), the heart (33) or the vasculature (34) have been widely reported, offering further support for the involvement of miRNAs in exercise adaptations. These studies have provided important pre-clinical evidence of potential therapeutic targets. Here, we will review miRNAs for their regulatory roles in physical fitness in these fields.
miRNAs in skeletal muscle adaption to exercise
The molecular basis of skeletal muscle adaptation to exercise is reflected by changes in contractile protein morphology and function (35), mitochondrial function (36), metabolic regulation, intracellular signaling and transcriptional responses including modification of MEF2, HDACs and NRFs. As the cellular composition of muscle, myofibers are the elementary units that make various adjustments after physical work to maintain optimal function including nerve stimulation, calcium signaling, and metabolic changes (37,38). Besides these well studied molecular mechanisms, experimental studies have identified fitness-related changes of miRNAs (39). In skeletal muscle, several miRNAs have been suggested to influence muscle myogenesis, muscle mass, and metabolism through their regulation of specific myogenic regulatory factors (MRFs) gene (40-42). Many are found to be dysregulated in skeletal muscle following physical training, which further supports the association of miRNAs with exercise-induced physiological changes and muscle function alteration (43). A more complete understanding of the miRNA biogenesis machinery following exercise in skeletal muscle may better guide the application of current therapies. Collectively, we summarize miRNAs function in muscle during physical training in the following sections.
miRNAs in skeletal muscle regeneration
Aging is associated with progressive changes in skeletal muscle mass and composition, which eventually results in the decline in muscle function, a process which universally affects patient mobility and quality of life. Exercise capacity is a well-known predictor of longevity. An upregulation of pri-miR-1 and pri-miR-133a has been identified in older men (44). Skeletal muscle expression of miR-1, which is responsible for muscle growth and satellite cell function, is elevated in young men but not older men after acute resistance exercise, demonstrating a divergent age-related muscle response after exercise. In addition, a novel role of the miR-126-IGF-1 axis has been identified in the control of exercise-induced adaptation of skeletal muscle, describing the dysregulation of this pathway in aging (45). Therefore, it appears that specific miRNAs may have an important role in the development of sarcopenia and, as such, may offer a source of novel therapeutics for the aging process.
Akt-mTOR signaling is attenuated during the onset and progression of age-related muscle wasting, however, it can be activated by resistance exercise (46). The miRNA species targeting the Akt-mTOR signaling have been investigated and it was found that 26 miRNAs were dysregulated with age and/or exercise (47). Among them, the miR-99/100 family of miRNAs notably emerged as potentially important regulators of Akt-mTOR signaling and muscle protein synthesis in response to resistance exercise in young and old subjects (47).
Taken together, these observations reveal how miRNAs, aging and exercise are interrelated and display the alteration of miRNAs by exercise may thus interfere with age-related muscle decline.
miRNAs in mitochondrial biogenesis
Physical exercise induces a range of signaling pathways, contributing to the metabolic and functional adaptations of skeletal muscle. PGC-1α is a key mediator of the transcriptional response in skeletal muscle to exercise, regulating mitochondrial metabolism, angiogenesis, β-oxidation and inflammation (48). Endurance exercise significantly increased the expression of miR-181, miR-1, and miR-107 and reduced miR-23 expression in C57Bl/6J wild-type male mice (49). miR-23-mediated post-transcriptional regulation of PGC-1α is potentially involved in the complex regulatory networks that govern skeletal muscle adaptation to endurance exercise (49). These findings of that study highlight the role of miRNAs in the control of PGC-1α-induced mitochondrial biogenesis in skeletal muscle post physical activity. Similarly, miR-696 was markedly downregulated by exercise targeting PGC-1α and regulated skeletal muscle biogenesis of mitochondria and fatty acid oxidation (50). In murine myoblast C2C12 cells during myogenic differentiation, the expression of miR-494 was markedly decreased, accompanied by an increase in mtDNA and the expression of predicted target genes for miR-494, including mitochondrial transcription factor A (mtTFA) and Forkhead box j3 (Foxj3), key transcription factors of mitochondrial biogenesis (51). Interestingly, miR-494 expression was significantly reduced in C57BL/6J mice skeletal muscle in endurance exercise, suggesting the vital role of miR-494 in exercise induced mitochondrial biogenesis via mtTFA and Foxj3 in vivo (51). The mRNA regulation of components of the miRNA biogenesis pathway (Drosha, Dicer and Exportin-5), muscle enriched miRNAs, (miR-1, miR-133a, miR-133b and miR-206), and several miRNAs dysregulated in muscle myopathies (miR-9, miR-23, miR-29, miR-31 and miR-181) was determined, and it was found that following an acute short-term exercise, the miRNA biogenesis pathway, as well as miR-1, miR-133a, miR-133b and miR-181a were all increased, while miR-9, miR-23a, miR-23b and miR-31 were decreased (52).
Expression of myomiRs, including miR-1, miR-133a, miR-133b and miR-206, were decreased after 12 weeks of endurance training in human muscle biopsies (53). Following 14 days after regular training, the levels myomiRs returned to baseline (53). Currently, the specific details of myomiRs directly alter human physiology in response to endurance exercise remains unknown. The details by which myomiRs alter human physiology in response to endurance exercise remain unknown.
However, it appears that exercise-induced miRNA alterations may be involved in the regulation of fundamental processes such as skeletal muscle regeneration, gene transcription and mitochondrial biogenesis, thereby contributing to skeletal muscle adaptation to exercise.
miRNAs in cardiovascular adaption to exercise
Exercise stimulates the hearts of athletes, resulting in physiological cardiac hypertrophy (54). In addition, exercise has been reported to provide sustainable protection against myocardial infarction (55) and ischemia-reperfusion injury (56). In contrast to pathological cardiac hypertrophy, which is featured with poor prognosis and heart failure (57), exercise-induced cardiac hypertrophy is generally accepted as protective, in some instances improves cardiac function, and does not progress to heart failure. This physiologic cardiac adaptation is characterized by an increase in heart size and cardiac output, resistance to and recovery from injury, and improved vascularization (3,33,57,58). Recently, miRNAs have received increasing attention in physical activity and cardiovascular health (59). The identification of miRNAs differentially regulated during physiological growth may open new therapeutic approaches for heart failure.
miRNAs in cardiac growth
Recently, miRNAs screening identified differentially expressed miRNAs that occur in response to physical training, and miR-222 was found to be upregulated in two distinct models of exercise (60). Circulating miR-222 increased in heart failure patients undergoing cardiopulmonary exercise testing. Inhibition of miR-222 by anti-miRs in vivo prevented exercise-induced cardiac growth. miR-222 cardiac overexpression mice were protected against ischemia-induced cardiac dysfunction and remodeling, implying a therapeutic cardioprotective role of miR-222 (61). Pathological left ventricular hypertrophy (LVH) is associated with increased local cardiac the renin angiotensin system (RAS) levels, represented by augmented angiotensinogen, angiotensin-converting enzyme (ACE) and angiotensin II (Ang II). Angiotensin-converting enzyme 2 (ACE2), a novel cardiac RAS, can maintains the important balance between the Ang II and Ang [1–7], favoring cardiovascular homeostasis. Swimming exercise training could decrease cardiac ACE and Ang II levels and increase ACE2 and Ang [1–7] levels (62). In addition, aerobic exercise training increased miR-27a and miR-27b, targeting ACE and decreased miR-143 targeting ACE2 in the heart, contributing to physiological LVH (62). miR-29 has also been reported to be decreased in cardiac hypertrophy induced by exercise in rat (63). Microarray analysis demonstrated downregulation of miR-1, miR-133a, and miR-133b and upregulation of miR-29a, however, only expression of miR-29a was associated with a significant decrease in LV collagen and an inverse correlation with hydroxyproline concentration indicating improved LV compliance and beneficial cardiac effects (63). The activation of PI3K/AKT/mTOR plays a critical role in the induction of physiological, but not pathological, cardiac hypertrophy (64-67). Recent work has suggested that the expression of miR-21, miR-144, and miR-145 were up-regulated in swimming exercise training-induced left ventricular remodeling while miR-124 was decreased (68). Moreover, they found that those miRNAs targeting the PI3K/AKT/mTOR signaling pathway and its negative regulators. A link between miRNAs and the PI3K/AKT/mTOR pathway has also been discovered in a rat model of cardiac hypertrophy induced by chronic swimming, in which miR-208a, miR-133b, miR-30e and miR-19b were upregulated, while miR-99b, miR-100, miR-191a, miR-22 and miR-181a were downregulated (69). Most of these miRNAs (e.g., miR-99, miR-100, miR-208, miR-181 and miR-19) were also associated with cardiac hypertrophy and apoptosis, principally acting via PI3K/Akt/mTOR and MAPK signaling pathways (69). A better understanding of miRNAs and the cellular pathways regulating exercise induced physiological cardiac hypertrophy may also assist in the development of treatments for CVD. Recently, a global analysis of miRNA expression in exercise-induced LVH identified an increase in miR-150 levels after 35 days and a decrease in miR-26b, miR-27a and miR-143 after 7 days of voluntary exercise (70). Further studies are needed to validate the targets of these miRNAs and to determine their functions in cardiac adaptation to physical training.
miRNAs in vascular remodeling
miR-126 and its validated targets, Sprouty-related protein 1 (Spred-1) and phosphoinositol-3 kinase regulatory subunit 2 (PI3KR2), were demonstrated to negatively regulate angiogenesis via the VEGF pathway inhibition in a rat model of swimming (71). In consist with it, hypertensive rats undertaking 10 weeks of swimming reduced blood pressure and heart rate, the expression of anti-angiogenic miRNAs miR-16 and miR-21 decreased in the soleus muscle, while miR-126 increased (72). Furthermore, physical activity increased angiogenic factors—VEGF, PI3KR2, and endothelial NO synthase (eNOS) in hypertension and inhibited apoptotic signaling (bcl2). Additionally, it was clearly indicated that exercise-induced angiogenic miRNAs influenced vascular disease, correcting capillary rarefaction and changes in fiber type distribution in hypertension and promoting a balance between angiogenic and apoptotic factors to prevent microvascular abnormalities in hypertension (72).
Circulating miRNAs (ci-miRNAs) adaption to exercise
Recently, several studies have demonstrated that miRNAs can be detected in the circulatory system and have been identified as a potential new class of biomarkers for health and disease (73-75). Mature miRNAs are released into body fluids by extracellular vesicles (76) or associated with RNA-binding proteins such as Argonaute2 (77) or lipoproteins such as HDL/LDL (78) secreted from various cell types including skeletal muscle (79). ci-miRNAs deliver the vesicle content to specific recipient target cells, exerting a paracrine function (80). It has been reported that exercise not only affects miRNA expression in the tissue but also in the circulation. ci-miRNAs can be easily sampled by body fluid and are stable following freezing, thawing, and temperature changes. Physical activity is associated with altered levels of ci-miRNAs, indicating that ci-miRNAs can be used as biomarkers to monitor such conditions (34).
To find exercise-associated alteration in ci-miRNAs expression in human, a profile of specific ci-miRNAs involved in angiogenesis (miR-20a, miR-210, miR-221, miR-222 and miR-328) (81), inflammation (miR-21 and miR-146a) (82), skeletal and cardiac muscle contractility (miR-21 and miR-133a) (83,84), and hypoxia/ischaemia adaptation (miR-21, miR-146a and miR-210) (85,86) have been analyzed in healthy competitive athletes before, during, and after acute exhaustive exercise testing. The levels of miR-20a, miR-21, miR-146a, miR-221 and miR-222 have been reported to be elevated in plasma after a 90-day period of aerobic exercise training. Among those miRNAs, they observed that peak levels of miR-146a and miR-20a positively correlated to peak oxygen consumption (VO2max), providing a feasible method of using ci-miRNAs as a biomarker of physical fitness (87). Later, if muscle-specific (miR-1, miR-133a, miR-133b, miR-208a, miR-208b, and miR-499) or muscle-related (miR-181 and miR-214) miRNAs were involved in physical activity was investigated. It was found that plasma levels of miR-1, miR-133a, and miR-133b were significantly increased 2 to 6 h after exercise, with a larger response following downhill training, while miR-181b and miR-214 were transiently up-regulated at the end of uphill exercise training (88). Other studies have been conducted to measure changes in muscle enriched ci-miRNAs in response to exercise. Circulating miR-486 (c-miR-486) was found to decrease after acute and chronic exercise, while other muscle enriched miRNAs such as miR-1, miR-16, miR-133a, miR-133b, miR-206, miR-208b and miR-499, have been difficult to measure due to low base-line expression levels. c-miR-486 displayed an inverse correlation with VO2max, suggesting that reduction of c-miR-486 mediates metabolic changes during physical exercise (89). In contrast with previous studies, another study evaluated the ci-miRNA profile after aerobic fitness and showed that the circulating levels of miR-149* increased, whereas miR-146a and miR-221 were significantly decreased. The microarray analysis revealed no changes in circulating levels of muscle specific miRNAs following acute resistance exercise. These differences imply that the changes in ci-miRNAs may be strongly influenced by exercise type, duration, and intensity (90).
To test the effect of physical work on endothelial cell damage, miR-126, as a marker for endothelial injury, was assessed by PCR analysis in plasma samples from healthy individuals following different endurance exercise tests, including maximal symptom-limited exercise test, bicycling, marathon, and resistance exercise. Circulating miR-126 increased in endurance exercise protocols as we mentioned above, providing evidence of endothelial damage caused by endurance exercise (91). Moreover, eccentric resistance training was found to cause increased levels of miR-133. Marathon completion leads to significant skeletal muscle damage, cardiac muscle stress/injury, and systemic inflammation, as reflected by statistically significant increases in circulating conventional biomarkers, including miRNAs. A cohort of miRNAs is involved in the dynamic regulation of ci-miRNAs before and after completion of prolonged, submaximal aerobic training (i.e., marathon run). ci-miRNAs enriched in muscle (miR-1, miR-133a, miR-499–5p) and cardiac tissue (miR-208a) display extremely low expression levels in plasma under resting conditions before marathon running, whereas ci-miRNAs enriched in the vascular endothelium (miR-126) and inflammatory miRNAs (miR-146a) are expressed at relatively higher levels (92). All candidate ci-miRNAs were found to be disproportionately increased immediately after the marathon and declined to pre-race levels or lower after 24 h of race completion. Another recent report of heart/muscle specific miRNAs in human before, directly after, and 24 h after a marathon run demonstrated a significant increase of miR-1, miR-133a, miR-206, miR-208b, and miR-499 (93). However, miR-499 and miR-208b returned to baseline, whereas the others were still raised 24 hours later. Significant correlations between VO2max, running speed at individual anaerobic lactate threshold (VIAS) and miR-1, miR-133a, miR-206, and the athlete’s aerobic performance capacity were found. In contrast, the expression of fibrosis/inflammation-associated miR-21 and miR-155 were not affected by exercise. Using a global ci-miRNA screen in young healthy men following an acute endurance exercise or 12-week endurance training, levels of ci-miRNAs including miR-338-3p, miR-330-3p, miR-223, miR-139-5p, miR-143 and miR-1 were found to be increased while the level of eight ci-miRNAs (miR-106a, miR-221, miR-30b, miR-151-5p, let-7i, miR-146a, miR-652 and miR-151-3p) decreased after acute exercise. Chronic alterations of ci-miRNAs in response to a 12-week endurance training have also been measured. Seven ci-miRNAs significantly decreased (miR-342-3p, let-7d, miR-766, miR-25, miR-148a, miR-185 and miR-21), while two ci-miRNAs significantly increased after the training period (miR-103 and miR-107) (94). As mentioned above, moderate aerobic exercise has been associated with strong anti-inflammatory mechanisms. The global response of circulating inflammation-related miRNAs (c-inflammamiRs) in response to different doses of acute exercise was investigated. The profiles of c-inflammamiRs before, immediately after, and 24 hours after participants ran 10-km and marathon races showed an increase in miR-150-5p immediately after the 10-km race while levels of 12 c-inflammamiRs were increased immediately after the marathon (let-7d-3p, let-7f-2-3p, miR-125b-5p, miR-132-3p, miR-143-3p, miR-148a-3p, miR-223-3p, miR-223-5p, miR-29a-3p, miR-34a-5p, miR-424-3p and miR-424-5p) (95). Taken together, these results indicate a clear dose-dependent effect of aerobic exercise on systemic inflammation and c-inflammamiR responses.
It has long been recognized that physical exercise has important benefits for diabetes. miR-192 and miR-193b have been identified as significantly increased in the prediabetic state but not in diabetic patients (96). Furthermore, circulating levels of miR-192 and miR-193b return to baseline in both prediabetic humans and glucose-intolerant mice undergoing chronic training, suggesting it may be a novel maker of pre-diabetes and a leading indicator for therapeutic exercise intervention.
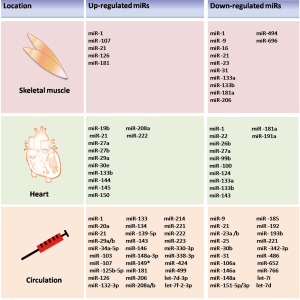
These results suggest the possibility of ci-miRNAs as an alternative, non-invasive and real-time circulating biomarker for exercise-induced tissue adaptation (Tables 1,2, Figure 2).
Table 1
| Ref. | Tissues | Model | Duration | MicroRNAs |
|---|---|---|---|---|
| Baggish et al., 2011 | Plasma | Rowing training | 90 days | miR-20a, -21, -146a, -221, -222↑ |
| Banzet et al., 2013 | Plasma | Walking exercises | 2/6/24/48/72 h | mir-1, -133a, -133b,-181b, -214, |
| Russell et al., 2013 | Muscle | Cycling exercise | 3 h | miR-1, -133a, -133-b, -181a,; miR-9, -23a, -23b, -31↓ |
| Short-term training | 10 days | miR-29be; miR-31↓ | ||
| Aoi et al., 2013 | Plasma | Cycling exercise | 3 h/24 h/4 weeks | miR-486↓ |
| Sawada et al., 2013 | Plasma | Bench/leg press | 0 h/1 h/1 day/3 days | miR-149*↑; miR-146a, -221↓ |
| Uhlemann et al., 2014 | Plasma | Maximal symptom limited exercise test | miR-126 | |
| Limited exercise test, bicycling | miR-126 | |||
| Marathon; resistance exercise | miR-126,-133e; miR-133, | |||
| Baggish et al., 2014 | Plasma | Marathon | 0/24 h | miR-1, miR-133a, -208a, -146a, -134, -126, 499–5p↑ |
| Mooren et al., 2014 | Plasma | Marathon | 0/24 h | miR-1, -133a, -206, -208b, -499↑ |
| Nielsen et al., 2014 | Plasma | Cycle ergometer exercise | 0/1/3 h | miR-338-3p, -330-3p, -223, -139-5p, -143, -1↑; miR-106a, -221, -30b, -151-5p, -146a, -652, -151-3p, let-7i6 |
| 12 weeks | miR-103, -107↑; miR-342-3p, -766, -25, -148a, -185, -21, let-7d↓ | |||
| de Gonzalo-Calvo et al., 2015 | Plasma | Marathon | let-7d-3p, let-7f-2-3p, miR-125b-5p, -132-3p, -143-3p, -148a-3p, miR-223-3p, -223-5p, -29a-3p,-34a-5p, -424-3p, -424-5p↑ | |
| Parrizas et al., 2015 | Plasma | miR-192, -193b9 | ||
| Rivas et al., 2014 | Muscle | Resistance exercise | miR-126n | |
| Safdar et al., 2009 | Muscle | Running | 3 h | miR-181, -1, -107↑; miR-230 |
| Aoi et al., 2010 | Muscle | Running | 4 weeks | miR-21g; miR-696I |
| Nielsen et al., 2010 | Muscle | Endurance training | 10 weeks | miR-1, -133a, -133b a, -206↓ |
| Fernandes et al., 2012 | Muscle | Swimming | 10 weeks | miR-126s; miR-16, -21↓ |
| D. A. Silva ND et al., 2012 | Muscle | Swimming | 10 weeks | miR-126s |
| Yamamoto et al., 2012 | Muscle | Swimming | 10 weeks | miR-494s |
| Russell et al., 2013 | Muscle | Endurance training | 3 h/10 days | miR-1, -133a, -133-b, -181a↑; miR-9, -23a, -23b, -31↓ |
| Liu et al., 2015 | Cardiac samples | Running or swimming | 3 weeks | miR-222 ↑ |
| Fernandes et al., 2011 | Cardiac samples | Swimming | 10 weeks | miR-27a,-27bl; miR-143, |
| Soci et al., 2011 | Cardiac samples | Swimming | 10 weeks | miR-29as; miRNAs-1, -133a, -133b↓ |
| Ma et al., 2013 | Cardiac samples | Swimming | 8 weeks | miR-21, -144, -145↑; miR-124 |
| Martinelli et al., 2014 | Cardiac samples | Metal wheels | 7/35 days | miR-150h; miR-26b, -27a, -143↓ |
| Ramasamy et al., 2015 | Cardiac samples | Swimming | 8 weeks | miR-208a,-133b,-30e,-19b1; miR-99b, -100, -191a, -22, -181a↓ |
Table 2
| MicroRNAs | Regulation | Target | Outcome | Ref. |
|---|---|---|---|---|
| miR-126 | Up | IGF-1 | Skeletal muscle regeneration | (45) |
| miR-99/100 | Up | Akt-mTOR signalling | Skeletal muscle regeneration | (47) |
| miR-23 | Up | PGC-1α | Skeletal muscle mitochondrial biogenesis | (49) |
| miR-696 | Up | PGC-1α | Skeletal muscle mitochondrial biogenesis | (50) |
| miR-494 | Up | PGC-1α | Skeletal muscle mitochondrial biogenesis | (51) |
| miR-222 | Up | P27/Hipk1/Hmbox1 | Cardiac hypertrophy | (62) |
| miR-27a/b | Up | ACE | Cardiac hypertrophy | (63) |
| miR-143 | Down | ACE2 | Anti-cardiac Hypertrophy | (63) |
| miR-21/miR-144 | Up | Pten | Cardiac hypertrophy | (69) |
| miR-145 | Up | TSC2 | Cardiac hypertrophy | (69) |
| miR-124 | Down | PIK3α | Anti-cardiac Hypertrophy | (69) |
| miR-126 | Up | Spred-1/ PI3KR2 | Angiogenesis | (72,73) |
| miR-16 | Down | VEGF /Bcl-2 | Anti-angiogenesis/apoptosis | (73) |
| miR-21 | Down | Bcl-2 | Apoptosis | (73) |
PIK3α, phosphoinositide-3-kinase catalytic alpha polypeptide; PTEN, phosphatase and tensin homolog; TSC2, tuberous sclerosis complex 2; Spred-1, sprouty-related protein 1; PI3KR2, phosphoinositol-3 kinase regulatory subunit 2; Hipk1, homeodomain interacting protein kinase 1;Hmbox1, homeobox containing 1.
lncRNAs and exercise
Although only a small number of functional lncRNAs have been well characterized to date, they can function via numerous paradigms and are key regulatory molecules in the cell. The discovery of lncRNAs highlights the rising interest in the roles of lncRNAs as a potentially new and crucial frontier for health and diseases. Recently, a conserved skeletal muscle-specific micropeptide, named myoregulin (MLN), encoded by a putative lncRNA, was discovered (97). MLN was embedded in the SR membrane, colocalized with SERCA1 and regulated Ca2+ handling by inhibiting SERCA pump activity. MLN KO mice showed improved exercise performance and Ca2+ handling in muscle. Those data linked the association of lncRNAs and skeletal muscle in physical training. Further studies to fully understand the role of lncRNAs for exercise will be of particularly interest.
Conclusions
Regular exercise directly alters skeletal muscle function, nutrient metabolism and muscle strength, along with reducing the risk of CVD, type 2 diabetes, and certain types of cancer. miRNAs have provided new insight into the understanding of the molecular mechanisms underlying exercise induced adaptations in skeletal muscle, the heart or the vasculature. Multiple studies in both animal models and humans suggest that miRNAs are dynamically regulated with physical activity. Unlike the well-known studies of miRNAs in fitness, little is known about lncRNAs in exercise training. The role of lncRNAs in exercise adaptation in skeletal muscle, the heart or the circulating system should be explored and developed further. Identifying the exercise-mediated signals regulating miRNAs and lncRNAs will be an important therapy and make physical work the most effective intervention to in the fight for the prevent disease development.
Acknowledgments
Funding: This work was supported by the grants from National Natural Science Foundation of China (81700351 and 31401238).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.09.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Roh J, Rhee J, Chaudhari V, et al. The role of exercise in cardiac aging: from physiology to molecular mechanisms. Circ Res 2016;118:279-95. [Crossref] [PubMed]
- Kraemer WJ, Fleck SJ, Evans WJ. Strength and power training: Physiological mechanisms of adaptation. Exerc Sport Sci Rev 1996;24:363-97. [Crossref] [PubMed]
- Sharma S, Merghani A, Mont L. Exercise and the heart: The good, the bad, and the ugly. Eur Heart J 2015;36:1445-53. [Crossref] [PubMed]
- Braith RW, Stewart KJ. Resistance exercise training: Its role in the prevention of cardiovascular disease. Circulation 2006;113:2642-50. [Crossref] [PubMed]
- Brown BM, Peiffer JJ, Martins RN. Multiple effects of physical activity on molecular and cognitive signs of brain aging: Can exercise slow neurodegeneration and delay alzheimer's disease? Mol Psychiatry 2013;18:864-74. [Crossref] [PubMed]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem 2006;75:19-37. [Crossref] [PubMed]
- Liu Y, Hu F, Li D, et al. Does physical activity reduce the risk of prostate cancer? A systematic review and meta-analysis. Eur Urol 2011;60:1029-44. [Crossref] [PubMed]
- Hawley JA, Hargreaves M, Joyner MJ, et al. Integrative biology of exercise. Cell 2014;159:738-49. [Crossref] [PubMed]
- Zierath JR, Wallberg-Henriksson H. Looking ahead perspective: Where will the future of exercise biology take us? Cell Metab 2015;22:25-30. [Crossref] [PubMed]
- Dangwal S, Schimmel K, Foinquinos A, et al. Noncoding RNAs in Heart Failure. Handb Exp Pharmacol 2017;243:423-45. [Crossref] [PubMed]
- Li J, Chan MC, Yu Y, et al. miR-29b contributes to multiple types of muscle atrophy. Nat Commun 2017;8:15201. [Crossref] [PubMed]
- Samanta S, Balasubramanian S, Rajasingh S, et al. MicroRNA: A new therapeutic strategy for cardiovascular diseases. Trends Cardiovasc Med 2016;26:407-19. [Crossref] [PubMed]
- Boon RA, Iekushi K, Lechner S, et al. Microrna-34a regulates cardiac ageing and function. Nature 2013;495:107-10. [Crossref] [PubMed]
- Su Z, Yang Z, Xu Y, et al. Micrornas in apoptosis, autophagy and necroptosis. Oncotarget 2015;6:8474-90. [Crossref] [PubMed]
- Lee Y, Jeon K, Lee JT, et al. Microrna maturation: Stepwise processing and subcellular localization. EMBO J 2002;21:4663-70. [Crossref] [PubMed]
- Lee Y, Kim M, Han J, et al. Microrna genes are transcribed by rna polymerase ii. EMBO J 2004;23:4051-60. [Crossref] [PubMed]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a rangtp-dependent dsrna-binding protein that mediates nuclear export of pre-mirnas. RNA 2004;10:185-91. [Crossref] [PubMed]
- Bartel DP. Micrornas: Target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny rnas with probable regulatory roles in caenorhabditis elegans. Science 2001;294:858-62. [Crossref] [PubMed]
- Sempere LF, Freemantle S, Pitha-Rowe I, et al. Expression profiling of mammalian micrornas uncovers a subset of brain-expressed micrornas with possible roles in murine and human neuronal differentiation. Genome Biol 2004;5:R13. [Crossref] [PubMed]
- McCarthy JJ. Microrna-206: The skeletal muscle-specific myomir. Biochim Biophys Acta 2008;1779:682-91.
- van Rooij E, Quiat D, Johnson BA, et al. A family of micrornas encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 2009;17:662-73. [Crossref] [PubMed]
- McCarthy JJ. The myomir network in skeletal muscle plasticity. Exerc Sport Sci Rev 2011;39:150-54. [Crossref] [PubMed]
- Kirby TJ, McCarthy JJ. Micrornas in skeletal muscle biology and exercise adaptation. Free Radic Biol Med 2013;64:95-105. [Crossref] [PubMed]
- Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding rna world. Oncotarget 2014;5:10976-96. [Crossref] [PubMed]
- Brosnan CA, Voinnet O. The long and the short of noncoding rnas. Curr Opin Cell Biol 2009;21:416-25. [Crossref] [PubMed]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding rnas. Mol Cell 2011;43:904-14. [Crossref] [PubMed]
- Gong C, Maquat LE. Lncrnas transactivate stau1-mediated mrna decay by duplexing with 3' utrs via alu elements. Nature 2011;470:284-88. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Srikantan S, et al. Lincrna-p21 suppresses target mrna translation. Mol Cell 2012;47:648-55. [Crossref] [PubMed]
- Ankö ML, Neugebauer KM. Long noncoding rnas add another layer to pre-mrna splicing regulation. Mol Cell 2010;39:833-34. [Crossref] [PubMed]
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding rna controls muscle differentiation by functioning as a competing endogenous rna. Cell 2011;147:358-69. [Crossref] [PubMed]
- Drummond MJ. Micrornas and exercise-induced skeletal muscle adaptations. J Physiol 2010;588:3849-50. [Crossref] [PubMed]
- Ellison GM, Waring CD, Vicinanza C, et al. Physiological cardiac remodelling in response to endurance exercise training: Cellular and molecular mechanisms. Heart 2012;98:5-10. [Crossref] [PubMed]
- Xu T, Liu Q, Yao J, et al. Circulating micrornas in response to exercise. Scand J Med Sci Sports 2015;25:e149-154. [Crossref] [PubMed]
- Widrick JJ, Stelzer JE, Shoepe TC, et al. Functional properties of human muscle fibers after short-term resistance exercise training. Am J Physiol Regul Integr Comp Physiol 2002;283:R408-16. [Crossref] [PubMed]
- Russell AP, Foletta VC, Snow RJ, et al. Skeletal muscle mitochondria: A major player in exercise, health and disease. Biochim Biophys Acta 2014;1840:1276-84.
- Rowe GC, Safdar A, Arany Z. Running forward: New frontiers in endurance exercise biology. Circulation 2014;129:798-810. [Crossref] [PubMed]
- Hoppeler H, Baum O, Lurman G, et al. Molecular mechanisms of muscle plasticity with exercise. Compr Physiol 2011;1:1383-412. [PubMed]
- Kirby TJ, Chaillou T, McCarthy JJ. The role of micrornas in skeletal muscle health and disease. Front Biosci (Landmark Ed) 2015;20:37-77. [Crossref] [PubMed]
- Rao PK, Kumar RM, Farkhondeh M, et al. Myogenic factors that regulate expression of muscle-specific micrornas. Proc Natl Acad Sci U S A 2006;103:8721-26. [Crossref] [PubMed]
- Güller I, Russell AP. Micrornas in skeletal muscle: Their role and regulation in development, disease and function. J Physiol 2010;588:4075-87. [Crossref] [PubMed]
- Rosenberg MI, Georges SA, Asawachaicharn A, et al. Myod inhibits fstl1 and utrn expression by inducing transcription of mir-206. J Cell Biol 2006;175:77-85. [Crossref] [PubMed]
- Aoi W. Frontier impact of micrornas in skeletal muscle research: A future perspective. Front Physiol. 2015;5:495. [Crossref] [PubMed]
- Drummond MJ, McCarthy JJ, Fry CS, et al. Aging differentially affects human skeletal muscle microrna expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab 2008;295:E1333-40. [Crossref] [PubMed]
- Rivas DA, Lessard SJ, Rice NP, et al. Diminished skeletal muscle microrna expression with aging is associated with attenuated muscle plasticity and inhibition of igf-1 signaling. FASEB J 2014;28:4133-47. [Crossref] [PubMed]
- Rommel C, Bodine SC, Clarke BA, et al. Mediation of igf-1-induced skeletal myotube hypertrophy by pi(3)k/akt/mtor and pi(3)k/akt/gsk3 pathways. Nat Cell Biol 2001;3:1009-13. [Crossref] [PubMed]
- Zacharewicz E, Della Gatta P, Reynolds J, et al. Identification of micrornas linked to regulators of muscle protein synthesis and regeneration in young and old skeletal muscle. PloS One 2014;9:e114009 [Crossref] [PubMed]
- Svensson K, Handschin C. Modulation of pgc-1alpha activity as a treatment for metabolic and muscle-related diseases. Drug Discov Today 2014;19:1024-29. [Crossref] [PubMed]
- Safdar A, Abadi A, Akhtar M, et al. Mirna in the regulation of skeletal muscle adaptation to acute endurance exercise in c57bl/6j male mice. PloS One 2009;4:e5610 [Crossref] [PubMed]
- Aoi W, Naito Y, Mizushima K, et al. The microrna mir-696 regulates pgc-1{alpha} in mouse skeletal muscle in response to physical activity. Am J Physiol Endocrinol Metab 2010;298:E799-806. [Crossref] [PubMed]
- Yamamoto H, Morino K, Nishio Y, et al. Microrna-494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcription factor a and forkhead box j3. Am J Physiol Endocrinol Metab 2012;303:E1419-27. [Crossref] [PubMed]
- Russell AP, Lamon S, Boon H, et al. Regulation of mirnas in human skeletal muscle following acute endurance exercise and short-term endurance training. J Physiol 2013;591:4637-53. [Crossref] [PubMed]
- Nielsen S, Scheele C, Yfanti C, et al. Muscle specific micrornas are regulated by endurance exercise in human skeletal muscle. J Physiol 2010;588:4029-37. [Crossref] [PubMed]
- Fagard R. Athlete’s heart. Heart 2003;89:1455-61. [Crossref] [PubMed]
- Hull SS Jr, Vanoli E, Adamson PB, et al. Exercise training confers anticipatory protection from sudden death during acute myocardial ischemia. Circulation 1994;89:548-52. [Crossref] [PubMed]
- Calvert JW, Condit ME, Aragon JP, et al. Exercise protects against myocardial ischemia-reperfusion injury via stimulation of beta(3)-adrenergic receptors and increased nitric oxide signaling: Role of nitrite and nitrosothiols. Circ Res 2011;108:1448-58. [Crossref] [PubMed]
- Hill JA, Olson EN. Cardiac plasticity. N Engl J Med 2008;358:1370-80. [Crossref] [PubMed]
- Hill JA. Braking bad hypertrophy. N Engl J Med 2015;372:2160-2. [Crossref] [PubMed]
- Tao L, Bei Y, Zhang H, et al. Exercise for the heart: Signaling pathways. Oncotarget 2015;6:20773-84. [Crossref] [PubMed]
- Liu X, Xiao J, Zhu H, et al. Mir-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab 2015;21:584-95. [Crossref] [PubMed]
- Uchida S, Dimmeler S. Exercise controls non-coding rnas. Cell Metab 2015;21:511-12. [Crossref] [PubMed]
- Fernandes T, Hashimoto NY, Magalhaes FC, et al. Aerobic exercise training-induced left ventricular hypertrophy involves regulatory micrornas, decreased angiotensin-converting enzyme-angiotensin ii, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1-7). Hypertension 2011;58:182-9. [Crossref] [PubMed]
- Soci UP, Fernandes T, Hashimoto NY, et al. Micrornas 29 are involved in the improvement of ventricular compliance promoted by aerobic exercise training in rats. Physiol Genomics 2011;43:665-73. [Crossref] [PubMed]
- Shioi T, Kang PM, Douglas PS, et al. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J 2000;19:2537-48. [Crossref] [PubMed]
- DeBosch B, Treskov I, Lupu TS, et al. Akt1 is required for physiological cardiac growth. Circulation 2006;113:2097-104. [Crossref] [PubMed]
- Condorelli G, Drusco A, Stassi G, et al. Akt induces enhanced myocardial contractility and cell size in vivo in transgenic mice. Proc Natl Acad Sci U S A 2002;99:12333-8. [Crossref] [PubMed]
- Oudit GY, Sun H, Kerfant BG, et al. The role of phosphoinositide-3 kinase and pten in cardiovascular physiology and disease. J Mol Cell Cardiol 2004;37:449-71. [Crossref] [PubMed]
- Ma Z, Qi J, Meng S, et al. Swimming exercise training-induced left ventricular hypertrophy involves micrornas and synergistic regulation of the pi3k/akt/mtor signaling pathway. Eur J Appl Physiol 2013;113:2473-86. [Crossref] [PubMed]
- Ramasamy S, Velmurugan G, Shanmugha Rajan K, et al. Mirnas with apoptosis regulating potential are differentially expressed in chronic exercise-induced physiologically hypertrophied hearts. PLoS One 2015;10:e0121401 [Crossref] [PubMed]
- Martinelli NC, Cohen CR, Santos KG, et al. An analysis of the global expression of micrornas in an experimental model of physiological left ventricular hypertrophy. PloS One 2014;9:e93271 [Crossref] [PubMed]
- DA Silva ND Jr. Swimming training in rats increases cardiac microrna-126 expression and angiogenesis. Med Sci Sports Exerc 2012;44:1453-62. [Crossref] [PubMed]
- Fernandes T, Magalhaes FC, Roque FR, et al. Exercise training prevents the microvascular rarefaction in hypertension balancing angiogenic and apoptotic factors: Role of micrornas-16, -21, and -126. Hypertension 2012;59:513-20. [Crossref] [PubMed]
- Li J, Xu J, Cheng Y, et al. Circulating microRNAs as mirrprs of acute coronary syndromes: MiRacle or quagMire? J Cell Mol Med 2013;17:1363-70. [Crossref] [PubMed]
- Creemers EE, Tijsen AJ, Pinto YM. Circulating micrornas: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 2012;110:483-95. [Crossref] [PubMed]
- Xu J, Zhao J, Evan G, et al. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J Mol Med (Berl) 2012;90:865-75. [Crossref] [PubMed]
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microrna. Nucleic Acids Res 2011;39:7223-33. [Crossref] [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating micrornas independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [Crossref] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. Micrornas are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Boon RA, Vickers KC. Intercellular transport of micrornas. Arterioscler Thromb Vasc Biol 2013;33:186-92. [Crossref] [PubMed]
- Vickers KC, Remaley AT. Lipid-based carriers of micrornas and intercellular communication. Curr Opin Lipidol 2012;23:91-7. [Crossref] [PubMed]
- Zhang C. Micrornas in vascular biology and vascular disease. J Cardiovasc Transl Res 2010;3:235-40. [Crossref] [PubMed]
- Davidson-Moncada J, Papavasiliou FN, Tam W. Micrornas of the immune system: Roles in inflammation and cancer. Ann N Y Acad Sci 2010;1183:183-94. [Crossref] [PubMed]
- Williams AH, Liu N, van Rooij E, et al. Microrna control of muscle development and disease. Curr Opin Cell Biol 2009;21:461-9. [Crossref] [PubMed]
- Davidsen PK, Gallagher IJ, Hartman JW, et al. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microrna expression. J Appl Physiol 2011;110:309-17. [Crossref] [PubMed]
- Thum T, Gross C, Fiedler J, et al. Microrna-21 contributes to myocardial disease by stimulating map kinase signalling in fibroblasts. Nature 2008;456:980-4. [Crossref] [PubMed]
- Chan SY, Loscalzo J. Microrna-210: A unique and pleiotropic hypoxamir. Cell Cycle 2010;9:1072-83. [Crossref] [PubMed]
- Baggish AL, Hale A, Weiner RB, et al. Dynamic regulation of circulating microrna during acute exhaustive exercise and sustained aerobic exercise training. J Physiol 2011;589:3983-94. [Crossref] [PubMed]
- Banzet S, Chennaoui M, Girard O, et al. Changes in circulating micrornas levels with exercise modality. J Appl Physiol 2013;115:1237-44. [Crossref] [PubMed]
- Aoi W, Ichikawa H, Mune K, et al. Muscle-enriched microrna mir-486 decreases in circulation in response to exercise in young men. Front Physiol 2013;4:80. [Crossref] [PubMed]
- Sawada S, Kon M, Wada S, et al. Profiling of circulating micrornas after a bout of acute resistance exercise in humans. PLoS One 2013;8:e70823 [Crossref] [PubMed]
- Uhlemann M, Mobius-Winkler S, Fikenzer S, et al. Circulating microrna-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol 2014;21:484-91. [Crossref] [PubMed]
- Baggish AL, Park J, Min PK, et al. Rapid upregulation and clearance of distinct circulating micrornas after prolonged aerobic exercise. J Appl Physiol 2014;116:522-31. [Crossref] [PubMed]
- Mooren FC, Viereck J, Kruger K, et al. Circulating micrornas as potential biomarkers of aerobic exercise capacity. Am J Physiol Heart Circ Physiol 2014;306:H557-63. [Crossref] [PubMed]
- Nielsen S, Akerstrom T, Rinnov A, et al. The mirna plasma signature in response to acute aerobic exercise and endurance training. PloS One 2014;9:e87308 [Crossref] [PubMed]
- De Gonzalo-Calvo D, Davalos A, Montero A, et al. Circulating inflammatory mirna signature in response to different doses of aerobic exercise. J Appl Physiol 2015;119:124-34. [Crossref] [PubMed]
- Párrizas M, Brugnara L, Esteban Y, et al. Circulating mir-192 and mir-193b are markers of prediabetes and are modulated by an exercise intervention. J Clin Endocrinol Metab 2015;100:E407-15. [Crossref] [PubMed]
- Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding rna regulates muscle performance. Cell 2015;160:595-606. [Crossref] [PubMed]
Cite this article as: Wang H, Liang Y, Li Y. Non-coding RNAs in exercise. Non-coding RNA Investig 2017;1:10.