
Anti-miR and gene therapy to heal the diseased heart
Cardiac remodelling leads to heart failure (HF) which represents a growing disease problem worldwide. Patients with HF have a high frequency of hospitalization and available pharmacological therapies are not sufficient to significantly reduce the high mortality. As standard care, physicians use potent blockers of e.g., single receptors in cardiac tissue thus impairing a single signalling cascade counteracting maladaptive cardiac deterioration. The identification of novel disease targets and the establishment of higher potency medication is thus an important aim in cardiovascular drug development. This strategy is supported by novel technologies sequencing whole genomes and highlighting the great majority of non-coding areas.
Since the last decade, post-transcriptional regulation via small non-coding microRNAs (miRNA, miR) has been reported in various HF conditions (1,2). The mode of action (MOA) for miRs is based on the repression of a set of target genes the so-called miRNA targetome, by way of example in cardiac ischemic conditions (2). First discovered in C. elegans (3,4), Watson-Crick mediated base pairing from nucleotides 2 to 8 (the seed sequence) of the miR and nucleotides of 3'-untranslated region (3'-UTR) in target genes mediate either mRNA degradation or translational inhibition. In line, single cardiac miRs like miR-34a and miR-132 are powerful regulators of cardiac performance and hold great promise for therapeutic modulation via antagonistic binding properties (5,6). Several clinical trials are already begun to study the effect of (mostly) anti-miR treatments in different HF settings (see clinicaltrials.gov) underlining the great potential of such pharmacological intervention.
To identify downstream miR effectors, researchers have to screen different bioinformatics tools available each with distinct feature or algorithm. Seed match between a miR and its targets as well as the conservation in seed region are the two major considerations contribute to the development of most miR target prediction tools, including the classical miRanda (7) and Targetscan (8). The stability of seed pairing sites is then estimated and scored based on the change in free energy (ΔG) since a negative ΔG indicates increased stability of system. Unlike most target prediction tools that accept only known miR and/or mRNA IDs, the plain sequences of miRs and the putative targets can also be utilized as input in RNA22 (9) and RNAhybrid (10), which provide the users a more free circumstance to evaluate any combination of miRNA:mRNA pairs. More recently, the accessibility of target region and the abundance of target sites in a given gene are also considered in tools such as PITA (11) and DIANA-microT-CDS (12). Although numerous miR targets prediction tools are available and easily applicable, cautions still need to be taken since each computational approach bears unique feature and limitation. Use of multiple prediction tools in parallel and carefully comparison of the different output is strongly recommended.
On the other hand, based on the sequencing of mRNA sites bound by the Argonaute (AGO) protein, high though put screen technology such as PAR-CLIP (13) improves the efficiency and the accuracy of experimentally miRNA target identification. Several publicly available databases incorporating large scale of experimentally validated miRNA-mRNA interactions e.g., Tarbase (14) have also been released. The increasing meta-data amount as well as the improved interface offered by public databases will tremendously facilitate the exploration of validated interaction between miRNAs and their targets.
In a recent issue of Circulation, Heggermont et al. (15) report that miR-146a and its downstream effector gene dihydrolipoyl succinyltransferase (DLST) are therapeutic targets in cardiac disease. Modelling of cardiac disease in a murine setting was accomplished applying transverse aortic constriction (TAC) or constant angiotensin-II (Ang-II) infusion via minipumps. Heggermont and colleagues identified enhanced miR-146a expression after disease stimulation and interestingly postulated an additional exosomal-based cell-to-cell communication pathway between endothelial cells and cardiomyocytes. To validate the importance of miR-146a abundancy for cardiac performance, Ang-II disease conditions were tested in mice with knockout for miR-146a (miR-146a ko). Of note, miR-146a ko animals were protected from the development of cardiac remodelling underlining a maladaptive function of elevated miR-146a expression levels. Mimicking the ko situation with a pharmacological intervention strategy applying small molecule blockers against miR-146a also demonstrated the effectiveness of blunted miR-146a expression to support cardiac healing. In contrast, cardiomyocyte-specific miR-146a overexpression led to cardiac hypertrophy already in resting conditions, thus response during cardiac stress was worsened. Mechanistically, several in silico and in vitro assays were performed to identify the metabolic factor dihydrolipoyl succinyltransferase (DLST) as a direct miR-146a target gene. Interestingly miR-146a has three potential binding sites in DLST 3'-UTR thereby mediating decay. Since DLST is a key factor within fatty acid and glucose metabolism, subsequent metabolome studies were performed in wildtype or miR-146a ko cardiomyocytes in vitro. Analysis clearly indicated an effect of lowered miR-146a expression on energy metabolism. Next to that, DLST protein expression was sustained in animals receiving anti-miR-146a highlighting an alternative approach to balance cardiac output. Using this observation nicely controlled studies were conducted applying a cardiotrophic adeno-associated virus serotype 9 (AAV9) to overexpress DLST in preventive manner. Early DLST overexpression could block the Ang-II induced effects on cardiac remodelling and impaired hypertrophic response.
Taken together, this combined study of (I) anti-miR-146a treatment and (II) the use of gene therapy to sustain the miR-146a target gene DLST suggest so-far unknown therapeutic approaches to improve functional cardiac parameters and to heal the diseased heart (Figure 1). These findings are of support to develop innovative miRNA-based therapeutics or AAV9-based vector systems in cardiovascular disease conditions. Accordingly, many open issues remain to address in future studies especially with respect towards the intrinsic translational potential. MOA in the current study is based on a targeted miR-146a-DLST-metabolome alteration axis in cardiomyocytes. The miR-146a inhibition strategy, however, is systemic thus other organs and cellular subtypes are affected where also MOA can be activated and presumably in an unwanted manner. To study potential off-target effects detailed studies (pharmacokinetics and pharmacodynamics) are needed to address such questions before completing translational evaluation. Taking this into account, site-specific delivery of anti-miR chemistry also calls for improvement, e.g. via the use of coupling oligonucleotides to antibody structures or encapsulating with activatable envelopes. In addition to the discovery of potent miR regulators, other cardiac non-coding RNAs alike long non-coding RNAs (16) or circularized RNAs (17), so-called circRNA, came into heavy research focus for the last years. To see which RNA subspecies makes up the winner in the race of novel innovative therapeutic drugs for HF is one of the great challenges in future cardiovascular medicine. Use of gene therapy alone, as it was stated for DLST in the herein discussed study, leaves enough space for discussion since it is unclear if this therapy is applicable in a human scenario where immune reaction is one of the biggest problems caused by synthetic gene supplementation.
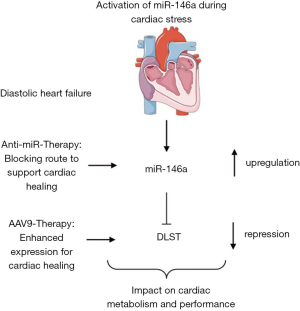
Despite aforementioned minor limitations, the study from Heggermont and co-workers highlights a novel strategy to target miR-146a and/or its downstream effector DLST to strengthen the diseased heart.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dongchao Lu (IFB, Molecular and Translational Therapeutic Strategies (Imtts), Hannover Medical School, Germany).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.09.04). TT reports support from the IFBTx (BMBF 01EO0802) and DFG TH 903/10-1; he has filed patents in the field of cardiovascular miRNA/lncRNA diagnostics and therapeutics; he's the founder and CSO of Cardior Pharmaceuticals GmbH. JF has filed patents in the field of cardiovascular miRNA/lncRNA diagnostics and therapeutics. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008;456:980-4. [Crossref] [PubMed]
- Fiedler J, Jazbutyte V, Kirchmaier BC, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 2011;124:720-30. [Crossref] [PubMed]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993;75:855-62. [Crossref] [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Ucar A, Gupta SK, Fiedler J, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012;3:1078. [Crossref] [PubMed]
- Boon RA, Iekushi K, Lechner S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013;495:107-10. [Crossref] [PubMed]
- Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol 2003;5:R1. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006;126:1203-17. [Crossref] [PubMed]
- Rehmsmeier M, Steffen P, Hochsmann M, et al. Fast and effective prediction of microRNA/target duplexes. RNA 2004;10:1507-17. [Crossref] [PubMed]
- Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet 2007;39:1278-84. [Crossref] [PubMed]
- Maragkakis M, Reczko M, Simossis VA, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 2009;37:W273-6 [Crossref] [PubMed]
- Hafner M, Landthaler M, Burger L, et al. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp 2010;2034 [PubMed]
- Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res 2015;43:D153-9. [Crossref] [PubMed]
- Heggermont WA, Papageorgiou AP, Quaegebeur A, et al. Inhibition of MicroRNA-146a and Overexpression of Its Target Dihydrolipoyl Succinyltransferase Protect Against Pressure Overload-Induced Cardiac Hypertrophy and Dysfunction. Circulation 2017;136:747-61. [Crossref] [PubMed]
- Fiedler J, Breckwoldt K, Remmele CW, et al. Development of Long Noncoding RNA-Based Strategies to Modulate Tissue Vascularization. J Am Coll Cardiol 2015;66:2005-15. [Crossref] [PubMed]
- Tan WL, Lim BT, Anene-Nzelu CG, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017;113:298-309. [PubMed]
Cite this article as: Fiedler J, Xiao K, Thum T. Anti-miR and gene therapy to heal the diseased heart. Non-coding RNA Investig 2017;1:12.


