
Looking for hints to mark functional lincs
It is generally accepted that long noncoding RNAs (lncRNAs) play fundamental roles in different physiological and pathological processes. However, out of the myriad of lncRNAs that have been detected, only a handful have been studied in sufficient detail to determine their function and the molecular mechanisms that support their function. This can be attributed to many reasons, among them the relative novelty of the lncRNA field and the numerous lncRNA genes to study. More importantly, the functional characterization of a particular lncRNA requires a considerable amount of resources, including time, money and technical expertise. It is fundamental to choose only those candidates that deserve such investment, which is not an easy task. At the beginning stages of lncRNA research many scientists will be confronted with long lists of lncRNAs that are, for instance, expressed or deregulated under a certain condition or bound by a specific factor. To select the best candidates for further analyses, it is much-needed to stablish a systematic way to prioritize lncRNAs according to functional traits. One of such efforts has been recently published by Marta Melé and colleagues in the paper entitled “Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs” (1).
LncRNAs are defined as transcripts longer than 200 nucleotides with poor capacity to translate for proteins (2). This modest description embraces a wide variety of RNAs generally shorter than a thousand nucleotides but with members that reach tens of thousands of nucleotides in length (3). Several scientists have striven to delve into the lncRNA jungle and sort non-coding transcripts under relevant categories. The most accepted classification of lncRNAs is based on their genomic location compared to neighboring genes. LncRNAs can be sense or antisense, when they overlap with one or more exons from another transcript in the same or in the opposite strand, respectively; intronic, when they overlap with an intron from another transcript; bidirectional or divergent, when they share the promoter with another transcript in the opposite strand; or intergenic (lincRNAs), when they are located between two genes. Interestingly, some lncRNAs function to regulate the expression of neighboring genes (4). Therefore, the genomic location of some lncRNAs may help to predict their function. When the function of a lncRNA has been determined, lncRNAs can be classified as cis and/or trans acting molecules. LncRNAs may act in trans, away from their site of transcription, or in cis, close to the site of synthesis. These cis-acting lncRNAs are naturally attached to the DNA and they may regulate the expression of genes located in a near locus or in the same nuclear territory (5). Certain cis-acting lncRNAs have been recently re-annotated as non-functional molecules with the discovery that it is not the lncRNA per se, but the sole act of transcription the one that drives functionality (6). Finally, transcriptional noise may give rise to some non-functional lncRNAs.
Few studies have classified lncRNA genes according to their epigenetic marks, transcription and processing. Instead, several authors have evaluated the similarities and differences between coding and noncoding genes and transcripts [reviewed in (7)]. LncRNAs and coding genes are very similar at chromatin level, where they share similar epigenetic marks, and at DNA level, where they show conserved transcription factor binding sites (TFBSs) (3,8,9). Similar to coding genes, most lncRNA genes are transcribed by polymerase II and processed by capping, splicing and, in ~40% of the cases, polyadenylation. Of note, lncRNAs tend to have fewer and longer introns than mRNAs and splicing is more inefficient. Compared to mRNAs, lncRNAs are less conserved, are expressed to lower levels, are more cell-type specific and locate more preferentially in the cell nucleus (3,10).
Melé and colleagues joined this line of research as they have compared several characteristics from a collection of 5,196 lncRNA genes and 19,575 coding genes. Interestingly, they have focused their analyses on lincRNAs to avoid overlapping regulation of coding genes and, for most experiments, only expression-matched groups of lincRNAs and mRNAs were used. After defining the promoter as the region located 5 kb upstream and downstream of the transcription start site (TSS), they analyzed 70 histone marks in seven ENCODE cell lines and 370 transcription factors (TFs) in eleven ENCODE cell lines (11). Tissue specificity was studied in RNA-seq data from 20 human tissues (8). Surprisingly, they have found that, compared to mRNA promoters, lincRNA promoters were depleted of almost all histone marks but H3K9me3 (histone H3 lysine 9 trimethylation) (Figure 1). H3K9me3 is a repressive mark bound by heterochromatin protein 1 (HP1), which can recruit repressive modifiers and contribute to heterochromatin compaction and spread (12). In fact, H3K9me3 silences noncoding repetitive regions. However, Melé and colleagues have found that the H3K9me3 mark does not correlate with deficient expression of lncRNAs. In fact, there is some evidence in the literature indicating that in certain cases, H3K9me3-enriched promoters may stimulate transcription initiation and elongation by RNA polymerase II (13,14). Unexpectedly, in five out of seven cell lines, promoters carrying the H3K9me3 mark were enriched in tissue-specific lincRNAs. Similar results were not observed when coding genes were used for the analysis. Further, compared to mRNA promoters, lincRNA promoters were bound by less TFs while enriched for certain families like GATA, JUN and FOS in all cell lines (Figure 1). Average conservation scores for TFBS showed a similar pattern: a general high score for lincRNA promoters that was significantly lower than that of mRNA promoters except for some specific cases like GATA2, KAP1 and MBD4, whose average conservation was higher in lincRNA promoters (Figure 1). Interestingly, expression of lincRNAs and coding genes increased with the number of conserved TFBSs, while tissue specificity decreased. The authors discuss that maintenance of basal mRNA expression across many tissues may require high numbers of TFBSs at the promoter of coding genes. LincRNA genes could generally be in a more repressed state that allows expression only in specific cells upon binding of a smaller set of TFs.
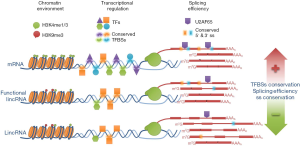
The authors have also investigated splicing efficiency in-depth. This emphasis makes particular sense in the case of lncRNAs because in spite of being less evolutionarily conserved than mRNAs, exceptional conservation has been reported when discrete regulatory motifs are analyzed, as has just been shown for TFBSs (15) and, very importantly, splicing consensus sequences (16,17). Using their own metric to calculate splicing efficiency (ratio of reads mapping to spliced isoforms versus total reads), Melé and colleagues have analyzed nuclear/cytosolic fractionated RNA-seq data from various cell lines. Similar to other studies, they have found that lncRNAs are much less efficiently spliced relative to expression matched mRNAs (18) (Figure 1). These results seem robust, as they were observed with nuclear transcripts and, although efficiencies were higher in general, with cytosolic transcripts as well. More so, results were reproduced in independent data sets with reliable annotations and with similarly fractionated RNA-seq data from mouse embryonic stem cells. Efficient splicing requires proper binding of U1 snRNP complex to the 5' splice site (ss) consensus sequence, binding of U2 snRNP and U2AF65 to the branch point and polypyrimidine track (PPT), a strong 3' ss consensus and the help of exonic splicing enhancers (ESE). In fact, the authors have found that ESE density is higher in lincRNAs than in mRNAs, probably because of the differences in GC content. However, the ESE density was not significantly different between efficient and inefficiently spliced lincRNAs, indicating that ESE density cannot explain the different splicing efficiency observed between lincRNAs and mRNAs. Similarly, the presence of U1 binding sites in the 5' ss of lincRNAs did not correlate significantly with the efficiency of splicing. Instead, the distance between the branch point and the 3' ss, a factor that correlates with splicing efficiency, was greater in lincRNAs than in mRNAs. In this region, the PPT of lincRNAs had less pyrimidines than the PPT of mRNAs and the number of pyrimidines correlated positively with splicing efficiency. In line with this finding, analysis of two independent CLIP-Seq data sets showed that binding of U2AF65 was depleted in lincRNAs compared to mRNAs (Figure 1). U2AF65 interacts with the PPT promoting the binding of U2 snRNP to the branch point. Therefore, the deficient splicing of lincRNAs could result from poor signals in the 3' region of the intron, where a generally shorter PPT may not support efficient U2AF65 binding.
The analyses of the splicing sequences performed in this and other studies, demonstrates that splicing of the general lincRNA population tends to be less efficient than that of mRNAs. Actually, the data presented by Melé and colleagues suggests that lincRNAs could be divided in those with efficient splicing (similar to that observed for mRNAs) and those with inefficient splicing. In this last group, splicing may not be required for functionality. The authors suggest that some of the lincRNAs that belong to this last group could be non-functional molecules such as AIRN, as it is not the lincRNA but the act of transcription the one that drives functionality (6). Similarly, non-functional RNAs generated from transcriptional noise could belong to the inefficiently-spliced group of lincRNAs. Instead, the group of efficiently spliced lncRNAs could represent functional molecules. In fact, several functional lncRNAs such as FIRRE, EGOT or NRIR have been shown to be efficiently spliced (19-21). Very importantly, to address whether this is the case in a larger scale, Melé and colleagues have studied the collection of functional lincRNAs deposited at the lncRNA database (lncRNAdb) (22). Indeed, the authors show that functional lincRNAs have higher splicing efficiency than non-functional transcripts.
Comparison of the features between lincRNAs included in the lncRNAdb and lincRNAs not functionally characterized has allowed the identification of certain traits that associate with functional lincRNAs: (I) higher splicing efficiency, (II) higher 5' and/or 3' ss conservation and (III) higher number of conserved TFBSs in the promoter (Figure 1). Then, functional lincRNAs are more similar to mRNAs than non-functional molecules. Interestingly, these criteria should be considered to select candidate lincRNAs for functional analyses.
Further studies are required to address whether the functional characteristics described for lincRNAs are general enough as to remain true for other subclasses of lncRNAs, including sense or antisense, intronic, bidirectional or divergent. In these cases, the interference of the coding sequences located nearby should be taken into account. Similar studies should also be applied to data generated by novel techniques, such as RACE-Seq, that aims to re-annotate lncRNA loci (23). Analyses of transcripts visualized by RACE-Seq indicate that about 60% of the genes targeted by the technique show 5' or 3' extensions and that the novel lncRNAs identified, are similar to mRNAs regarding their length and the number of exons and alternatively spliced isoforms. These datasets provide a solid resource to determine splicing efficiency, ss conservation and to identify more accurately lncRNA promoters.
In conclusion, it is of paramount importance for the field to determine key factors that help to separate functional lncRNAs from the noisy transcription plaguing eukaryotic genomes. In the meantime, the study performed by Melé and colleagues is more than a fairly thorough characterization of both cis- and trans-acting elements regulating lincRNA biogenesis. The inquiry for enrichment from a pool of functional lncRNAs should become standard for similar future studies. The criteria that they have described to tackle functional lincRNAs may be already useful at the beginning stages of lncRNA research and may serve as a solid base for other studies aiming to define factors with enough discriminatory power to mark functional lncRNAs.
Acknowledgments
Funding: Spanish Department of Science (SAF2015-70971-R), grant Ortiz de Landazuri from the Government of Navarra and European FEDER funding. JPU is a recipient of a fellowship from the University of Navarra’s Asociación de Amigos.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Meiyi Song (Division of Gastroenterology and Hepatology, Digestive Disease Institute, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.11.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Melé M, Mattioli K, Mallard W, et al. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res 2017;27:27-37. [Crossref] [PubMed]
- Garitano-Trojaola A, Agirre X, Prósper F, et al. Long non-coding RNAs in haematological malignancies. Int J Mol Sci 2013;14:15386-422. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Barriocanal M, Carnero E, Segura V, et al. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Front Immunol 2015;5:655. [Crossref] [PubMed]
- Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol 2012;19:1068-75. [Crossref] [PubMed]
- Latos PA, Pauler FM, Koerner MV, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012;338:1469-72. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223-7. [Crossref] [PubMed]
- Necsulea A, Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet 2014;15:734-48. [Crossref] [PubMed]
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004;306:636-40. [Crossref] [PubMed]
- Becker JS, Nicetto D, Zaret KS. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet 2016;32:29-41. [Crossref] [PubMed]
- Lee S, Lee J, Chae S, et al. Multi-dimensional histone methylations for coordinated regulation of gene expression under hypoxia. Nucleic Acids Res 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Wiencke JK, Zheng S, Morrison Z, et al. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene 2008;27:2412-21. [Crossref] [PubMed]
- Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014;505:635-40. [Crossref] [PubMed]
- Haerty W, Ponting CP. Unexpected selection to retain high GC content and splicing enhancers within exons of multiexonic lncRNA loci. RNA 2015;21:333-46. [Crossref] [PubMed]
- Nitsche A, Rose D, Fasold M, et al. Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 2015;21:801-12. [Crossref] [PubMed]
- Eser P, Wachutka L, Maier KC, et al. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. Mol Syst Biol 2016;12:857. [Crossref] [PubMed]
- Kambara H, Niazi F, Kostadinova L, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res 2014;42:10668-80. [Crossref] [PubMed]
- Hacisuleyman E, Goff LA, Trapnell C, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 2014;21:198-206. [Crossref] [PubMed]
- Carnero E, Barriocanal M, Prior C, et al. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 2016;17:1013-28. [Crossref] [PubMed]
- Amaral PP, Clark MB, Gascoigne DK, et al. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 2011;39:D146-51. [Crossref] [PubMed]
- Lagarde J, Uszczynska-Ratajczak B, Santoyo-Lopez J, et al. Extension of human lncRNA transcripts by RACE coupled with long-read high-throughput sequencing (RACE-Seq). Nat Commun 2016;7:12339. [Crossref] [PubMed]
Cite this article as: Unfried JP, Fortes P. Looking for hints to mark functional lincs. Non-coding RNA Investig 2017;1:17.


