
The non-coding genome in cancer
The completion of the Human Genome Project in 2003 led to the launch of several major projects, including the international HapMap Project to identify genetic variants and haplotypes in the human genome (1), the 1000 Genomes Project to characterize the frequency of genetic variants in human populations (2), the ENCODE project to identify functional elements in the human genome (3,4), and the ROADMAP project to assess epigenetic alternation of DNA sequences (5). All these projects have yielded unprecedented information on the human genome: for instance, exon regions of genes are seen to make up less than 2% of the human genome. Most of the human genome (98%) is thus non-coding but contains many regulatory elements, including enhancers, silencers, insulators, or locus control regions (LCR).
The non-coding regulatory regions of the human genome have been found to be enriched for DNase I hypersensitive sites (DHS), histone modification regions, DNA methylation regions, and transcription factor binding sites (6,7). In recent years, up to 75% of the human genome was also observed to be transcribed, generating thousands of non-coding RNAs (8), of which long non-coding RNAs (lncRNAs) represent the largest group (9). Increasing evidence shows that lncRNAs may regulate gene expression via diverse biological mechanisms, such as epigenetic regulation, chromatin remodeling, and gene transcription, but they may also play a role in cellular transport, metabolic processes, and chromosome dynamics (10). Many lncRNAs have been linked to disease phenotypes, for example, a liver-specific lncRNA LIVAR was reported to affect hepatocyte viability and its expression level was associated with non-alcoholic fatty liver disease (NAFLD), suggesting it has a protective effect in NAFLD (11).
The importance of non-coding regions in health and disease has been demonstrated by genome-wide association studies (GWAS) and the vast majority (about 93%) of the reported genetic variants lie in non-coding regions and are enriched for regulatory regions, like enhancers and DHS regions. These non-coding variants are also enriched for eQTL effects and affect the expression of both protein-coding genes and non-coding RNAs (12). Linking non-coding variants to functional consequences can yield mechanistic insights into disease mechanisms. Two examples are: (I) a candidate causal SNP was predicted to alter RNUX transcription factor binding in regulatory regions relevant to breast cancer, thereby affecting expression of its downstream genes (13) and (II) GWAS variants linked to atherosclerosis-related phenotypes were associated with a lower expression of lncRNA ANRIL, the knock-down of which leads to reduced cell growth, possibly via CDKN2A/B regulation (14).
In addition to large numbers of non-coding germ line variants, the vast majority of somatic mutations in cancer genomes occur in non-coding regions (15), although previous cancer genomics studies have focused on coding regions. For instance, The Cancer Genome Atlas (TCGA) reported somatic mutations in 3,281 tumors across 12 major cancer types using whole exon sequencing (16), while more recently, the Memorial Sloan Kettering (MSK) Cancer Center identified genetic mutations in more than 10,000 cancer patients using hybridization captured-based NGS panel (MSK-IMPACT), which captures only a small number of non-coding sites (17). However, there is increasing interest in the role of non-coding variants in cancer (15,18). The somatic mutations in non-coding regions are believed to promote tumorigenesis, together with mutations in coding regions. However, very few non-coding drivers have been identified so far and cancer mutations in non-coding regions are poorly characterized.
Li et al. recently reported on ‘Whole-genome analysis of papillary kidney cancer finds significant non-coding alterations’ (PLoS Genet 2017) investigating the impact of non-coding alterations in one of the most common kidney cancer (19). Li et al.’s work adds to the many new gene mutations that have been linked to papillary renal cell carcinoma (pRCC), although the driver genes and pathways are still unknown in many cases. They aimed to explore the potential non-coding drivers and heterogeneity of the cancer by performing the first whole-genome sequencing analysis on tumor samples from 35 pRCC patients. First, they focused on MET (tyrosine kinase), a known driver gene in pRCC. In the non-coding regions of MET, they found that a cryptic promoter in the second intron initiates expression of a pRCC-associated alternative transcript. Using a methylation array probe, a significantly lower methylation level was seen in samples expressing the alternative transcript, suggesting that methylation changes may drive pRCC development via MET. Moreover, Li et al. reported mutations in the MET promoter and in the first two introns where the alternative splicing starts. However, they did not find any correlation between the alternative splicing events and intronic mutations, so this needs further investigation. Next they evaluated other non-coding regions throughout the genome. A mutation hotspot on chromosome 1 was detected in 6 out of 35 samples. This hotspot overlapped the predicted regulatory region at the 5’ end of ERRFI1 (ERBB receptor feedback inhibitor 1), a negative regulator of the cancer-associated genes EGFR, HER2 and HER3. Hence ERRFI1 may serve as potential tumor suppressor. However, no changes in mRNA, protein or phosphorylation levels of these proteins were observed, but this might be due to the limited sample size. Another hotspot was observed in a putative promoter and flanking region of NEAT1, a cancer-associated lncRNA. These mutations were associated with higher mRNA levels of NEAT1 and with a worse prognosis for the patient. Mutations in NEAT1 have also been reported in other cancer studies (20,21). NEAT1 mRNA expression was highly correlated with expression levels of the downstream gene MALAT1, another lncRNA associated with cancer (22). These two lncRNAs may use a similar mechanism to regulate cancer progression. Furthermore, from their whole genome sequencing analysis of 35 pRCC samples, Li et al. identified some interesting characteristics of somatic mutation spectra. The mutations in pRCC patients were enriched for C-to-T transmission at CpG sites, which was associated with a lower methylation level. However, these mutations were enriched for coding regions and were non-synonymous. Interestingly, the mutations in DHS sites are likely driven by defects in chromatin remodeling as the authors showed that defects in chromatin remodeling genes could result in a 60% increase in the number of mutations in DHS regions. The potential mechanism of how DHS mutations could affect gene transcription is presented in Figure 1. Over 95% of DHS sites are positioned distally from exons regions, with half in intronic regions and half in intergenic regions (23). This implies that mutations in coding regions can result in somatic mutations in non-coding regions.
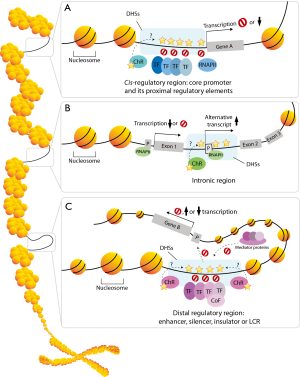
Li et al. have shown that non-coding alteration is common in pRCC patients and they have characterized mutation spectra at the whole genome level and DHS sites. However, it remains unclear whether these non-coding variants are only errors due to defects in DNA repairs, as shown by the defects in chromatin remodeling genes. It would be interesting to investigate how probable somatic non-coding mutations can contribute to tumorigenesis. And it is certainly important to further investigate the functional effects of the somatic non-coding mutations and relate them to the results from various omics profiling, next-generation sequencing technologies like ChIP-seq and RNA-Seq, and state-of-the-art molecular techniques such as CRISPR-Cas genome editing.
Acknowledgments
We thank Jackie Senior for editing the manuscript.
Funding: This work was supported by grants from the Jan Kornelis de Cock Foundation (to B Atanasovska); the Netherlands Organization for Scientific Research (NWO-VIDI 864.13.013 to J Fu); the Systems Biology Center for Metabolism and Ageing, Groningen, the Netherlands (SBC-EMA to J Fu); the BBMRI-NL complementation project (to J Fu); and CardioVasculair Onderzoek Nederland (CVON 2012-03 to J Fu).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Meiyi Song (Division of Gastroenterology and Hepatology, Digestive Disease Institute, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.12.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- International HapMap Consortium. The International HapMap Project. Nature 2003;426:789-96. [Crossref] [PubMed]
- 1000 Genomes Project Consortium, Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061-73.
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004;306:636-40. [Crossref] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [Crossref] [PubMed]
- Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317-30. [Crossref] [PubMed]
- Lee D, Gorkin DU, Baker M, et al. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet 2015;47:955-61. [Crossref] [PubMed]
- Zeng H, Gifford DK. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res 2017;45:e99 [Crossref] [PubMed]
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalogue of human long non-coding RNAs: Analysis of their structure, evolution and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Devaux Y, Zangrando J, Schroen B, et al. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol 2015;12:415-25. [Crossref] [PubMed]
- Atanasovska B, Rensen SS, van der Sijde MR, et al. A liver-specific long noncoding RNA with a role in cell viability is elevated in human nonalcoholic steatohepatitis. Hepatology 2017;66:794-808. [Crossref] [PubMed]
- Degner JF, Pai AA, Pique-Regi R, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 2012;482:390-4. [Crossref] [PubMed]
- Liu Y, Walavalkar NM, Dozmorov MG, et al. Identification of breast cancer associated variants that modulate transcription factor binding. PLoS Genet 2017;13:e1006761 [Crossref] [PubMed]
- Congrains A, Kamide K, Oguro R, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012;220:449-55. [Crossref] [PubMed]
- Khurana E, Fu Y, Chakravarty D, et al. Role of non-coding sequence variants in cancer. Nat Rev Genet 2016;17:93-108. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. [Crossref] [PubMed]
- Weinhold N, Jacobsen A, Schultz N, et al. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 2014;46:1160-5. [Crossref] [PubMed]
- Li S, Shuch BM, Gerstein MB. Whole-genome analysis of papillary kidney cancer finds significant noncoding alterations. PLoS Genet 2017;13:e1006685 [Crossref] [PubMed]
- Li Y, Li Y, Chen W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget 2015;6:27641-50. [PubMed]
- He C, Jiang B, Ma J, et al. Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. APMIS 2016;124:169-74. [Crossref] [PubMed]
- Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res 2015;75:1322-31. [Crossref] [PubMed]
- Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature 2012;489:75-82. [Crossref] [PubMed]
Cite this article as: Atanasovska B, Fu J. The non-coding genome in cancer. Non-coding RNA Investig 2018;2:4.


