
Lnc-ing noncoding RNAs to Wnt signaling in colorectal cancer
Large-scale transcriptomic studies have shown that the majority of the genome is transcribed, however, only a small proportion codes for protein (1). The majority of noncoding transcripts produced are long noncoding RNAs (lncRNAs), loosely defined as transcripts >200 nucleotides that do not code for protein. Similar to mRNAs, the majority of lncRNAs are transcribed by RNA polymerase II and spliced using consensus splice sites (2). The exact proportion of noncoding transcripts that represent functional genes has been a hot topic for debate. However, there is no doubt that many of these transcripts are indeed functional, are key regulators of biological processes and act through an increasingly diverse array of mechanisms (3).
Multiple lncRNAs have now been shown to play important roles in cancer development and metastasis, including colorectal cancer (CRC). Notable examples include HOTAIR (HOX transcript antisense RNA), with high HOTAIR expression highly correlated with liver metastases and poor prognosis in CRC patients (4). Several studies have shown that HOTAIR promotes epithelial to mesenchymal transition (EMT) by directly binding the polycomb repressive complex 2 (PRC2) to reprogram PRC2 occupancy (5). Colon cancer associated transcript 1 (CCAT1) expression, a lncRNA transcribed from a super enhancer at 8q24, is associated with poor prognosis and can predict patient response to bromodomain and extra-terminal motif (BET) inhibitors (6). Another example is N-BLR, a primate-specific lncRNA, which promotes EMT by directly interacting with miR-141 and miR-200c-3p and acting as a miRNA sponge (7).
The recent study by Lin et al. (8), provides another example of a lncRNA involved in CRC invasion and metastasis. The authors first profiled lncRNA expression by microarray in six human CRC and normal adjacent tissue. They identified 2,545 lncRNAs differentially expressed between the CRC and normal samples, of which five were further validated by qPCR. Specifically, they showed that lncRNA BC032913 was markedly down regulated in CRC tissues, and associated with an advanced tumor stage, distant metastases and poor patient survival, suggesting that it may participate in tumor metastasis.
Lin and colleagues then investigated the function of BC032913 through a combination of in vitro assays (8). They found ectopic expression of BC032913 inhibited cell migration and invasion in HCT116 and DLD-1 cells. To further investigate the phenotype, the authors profiled metastasis-related genes in HCT116-control versus -BC032913 cells using a human tumor metastasis PCR array. They found BC032913 significantly increased the expression of several genes, including TIMP3 (TIMP metallopeptidase inhibitor 3), a secreted protein that inhibits extracellular matrix remodelling (9). This result was further confirmed by Western blot in HCT116-BC03213 and DLD1-BC03213 cells that showed a concomitant increase in TIMP3 protein levels.
Several studies report that TIMP proteins can promote tumorigenesis through aberrant regulation of the Wnt/β-catenin pathway (10,11). Given the importance of Wnt/β-catenin signaling in CRC (12), the authors hypothesized that increased BC032913-TIMP3 levels may alter this key pathway. To explore this connection, Lin et al. used TOPflash reporter assays to show that BC0321913 overexpression significantly reduced luciferase activity, and that this effect was partially rescued by TIMP3 silencing. Western blot also revealed that BC0321913 increased E-cadherin levels, but downregulated nuclear β-catenin and CD44, which could be reversed by TIMP3 silencing. CD44 is a downstream Wnt target gene and has previously been reported to promote EMT in CRC (13).
Finally, the authors investigated the role and potential mechanism of BC032913 on CRC metastasis in vivo. They subcutaneously injected HCT116-control or -BC032913 into the tail vein or spleens of nude mice and observed fewer and smaller micrometastases in the HCT116-BC032913 group. They also analysed the expression of TIMP3 and β-catenin by qPCR in metastases excised from individual mice in each group. Consistent with the in vitro data, the mRNA levels for TIMP3 and β-catenin were increased and decreased, respectively. While not conclusive, collectively the data presented does provide evidence that BC032913 may directly or indirectly inhibit CRC metastasis via inactivation of the Wnt/β-catenin pathway with subsequent effects on EMT (Figure 1).
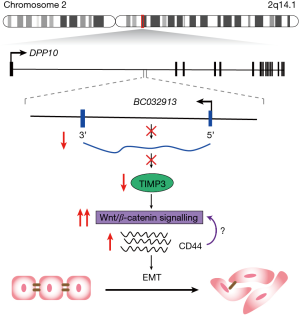
The exact mechanism by which BC032913 mediates its actions is unclear. Of note, BC032913 is antisense to intron 2 of dipeptidyl peptidase like 10 (DPP10) and shares a bidirectional promoter with an alternative isoform of DPP10 (DPP10-2). Previous studies have shown that, in a subset of neurons, BC032913 (also known as LOC389023) acts as a transcriptional repressor of DPP10-2, likely through the recruitment of components of the PRC2 complex (14). Lin et al. indicated that DPP10 mRNA levels were decreased in CRC tissue however, DPP10 expression could not be detected in a panel of CRC cell lines making it difficult to assess the relationship between DPP10 and BC032913. Future studies will be required to determine whether BC032913 is chromatin associated in colorectal cells similar to what has been observed in neurons. Identifying proteins or miRNAs that interact with the lncRNA will also provide important insights into the mechanism by which BC032913 promotes cancer progression.
In summary, Lin et al. have characterised a novel lncRNA in CRC metastasis. This lncRNA now adds to an ever-increasing list of noncoding RNAs involved in cancer progression. The authors suggest that BC032913 may serve as a new biomarker or therapeutic target for CRC. This may prove difficult given that BC032913 is down-regulated in CRC however, targeting key regulators of BC032913 may provide an alternative avenue for boosting the levels BC032913 in CRC cells.
Acknowledgments
Funding: This work was supported by a grant from the National Health and Medical Research Council of Australia (NHMRC; APP1122022). An NHMRC Senior Research Fellowship (APP1135932) supported SLE.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Jinzhe Zhou (Department of General Surgery, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.01.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Hon CC, Ramilowski JA, Harshbarger J, et al. An atlas of human long non-coding RNAs with accurate 5' ends. Nature 2017;543:199-204. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med 2015;12:1-9. [PubMed]
- McCleland ML, Mesh K, Lorenzana E, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Invest 2016;126:639-52. [Crossref] [PubMed]
- Rigoutsos I, Lee SK, Nam SY, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol 2017;18:98. [Crossref] [PubMed]
- Lin J, Tan X, Qiu L, et al. Long Noncoding RNA BC032913 as a Novel Therapeutic Target for Colorectal Cancer that Suppresses Metastasis by Upregulating TIMP3. Mol Ther Nucleic Acids 2017;8:469-81. [Crossref] [PubMed]
- Arpino V, Brock M, Gill SE. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol 2015;44-46:247-54. [Crossref] [PubMed]
- Xia Y, Wu S. Tissue inhibitor of metalloproteinase 2 inhibits activation of the beta-catenin signaling in melanoma cells. Cell Cycle 2015;14:1666-74. [Crossref] [PubMed]
- Blavier L, Lazaryev A, Dorey F, et al. Matrix metalloproteinases play an active role in Wnt1-induced mammary tumorigenesis. Cancer Res 2006;66:2691-9. [Crossref] [PubMed]
- White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/beta-catenin signaling in gastrointestinal cancers. Gastroenterology 2012;142:219-32. [Crossref] [PubMed]
- Cho SH, Park YS, Kim HJ, et al. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int J Oncol 2012;41:211-8. [PubMed]
- Shulha HP, Crisci JL, Reshetov D, et al. Human-specific histone methylation signatures at transcription start sites in prefrontal neurons. PLoS Biol 2012;10:e1001427 [Crossref] [PubMed]
Cite this article as: Edwards SL, French JD. Lnc-ing noncoding RNAs to Wnt signaling in colorectal cancer. Non-coding RNA Investig 2018;2:7.


