
Long intergenic non-coding RNAs in hepatocellular carcinoma—a focus on Linc00176
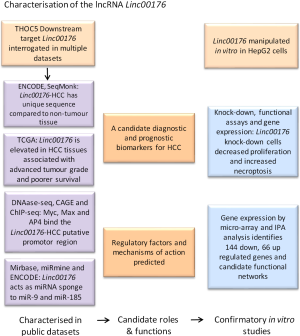
Liver cancer is a common type of cancer that ranks the second in cancer-related mortality world-wide (1). Hepatocellular carcinoma (HCC) is the predominant type of liver cancer and it usually arises on the background of cirrhosis. Risk factors associated with the incidence of HCC include viral hepatitis (hepatitis B and C viruses), the metabolic syndrome including non-alcoholic fatty liver diseases, autoimmune hepatitis as well as aflatoxin-B ingestion (2,3). The multikinase inhibitor, sorafenib, is the only 1st line medical therapy widely available, with its sister drug regorafenib approved for 2nd line use in selected patients (4,5). These drugs have only a limited impact on life expectancy. Transcriptomic changes accompanying cancer development and progression have been extensively explored in patients with HCC, with subgroups created based on transcription profile and mutational status (6-9). These studies to date have not adequately informed selection of candidate drugs for clinical trials, the vast majority of which have failed to show any survival benefit. Although trial design and toxicities have contributed to failures (8), our lack of global understanding of the tumour biology is likely also to have played a role. Recently, rather than sticking to the central transcriptome dogma of DNA-mRNA-protein, researchers have started to appreciate the many other transcripts out of this workflow that are cancer specific and functional, as alternative sources for the identification of novel candidate therapeutic targets. High-throughput sequencing techniques in combination with advanced computational predicting software and epigenetic tools have identified novel RNA transcripts and are even capable of predicting specific functions. These novel identified transcripts include “long non-coding RNAs (lncRNAs)”, a term generating more than 13,500 hits in a PubMed search at the end of 2017—three quarters of which were published in the last decade. These include a recent article by Tran and colleagues, published in Oncogene, in which the lncRNA Linc00176 has been interrogated in publicly available datasets. It has been reported as an upregulated transcript in HCC that is worthy of pursuit as a biomarker directed target for cancer therapy (10).
The term lncRNA defines a category of RNA transcripts more than 200 nucleotides in length that don’t encode for proteins. These features differentiate lncRNAs from short transcripts like miRNAs, tRNAs or snoRNAs, as well as from mRNAs. LncRNA and mRNA share similarities in the regulation of their transcription and post-transcriptional processing, but lncRNAs are shorter in length and have fewer exons and conserved primary structures compared to protein coding transcripts (11). Integration of the human RNA transcriptome with advanced bioinformatics analysis has indicated that over 60% of transcripts in the so called “MiTranscriptome” of human long poly-adenylated RNA transcripts are lncRNAs (12). Located within the intergenic regions and expressed at lower levels than protein coding RNAs, unsupervised clustering of the differentially expressed transcripts has identified the existence of distinct tissue-specific signatures of lncRNAs. Moreover, lncRNAs are differentially expressed between cancer and normal tissue within the same organ (12). In the liver, RNA sequencing of 60 primary HCC in Chinese patients, matched with tissue from portal vein tumour thrombosis (PVTT) and adjacent non-tumour liver, has identified a deregulated pool of lncRNAs in the primary and metastatic tumours (13). Remarkably, more than 75% of sequenced lncRNAs identified in HCC had not been previously annotated in either the MiTranscriptome (12) or the GENCODE transcriptome (14). In this cohort of Chinese patients with HCC, DNA methylation and copy number variation (CNV) were associated with deregulated lncRNAs pool and the lncRNA transcripts appeared to have regulatory roles in cellular functions, such as the immune response and cell adhesion. The tissue and cancer specificity of lncRNAs, alongside key regulatory roles, implies importance in cancer pathophysiology and highlights their potential as novel cancer-specific targets.
The transcription/export (TREX) mRNA export complex is a master regulator of mRNA biogenesis, being involved in different steps of mRNA transcription, processing and export. The mammalian TREX complex includes THOC1 (hHpr1), THOC2, THOC7, THOC5 (FMIP), THOC6, THOC3 (hTEX1), Uap56, DDX39c and Aly (15). The Hannover research group led by Teruko Tamura has previously shown that THOC5 null mice have drastically reduced numbers of hematopoietic system and myeloid progenitor cells, without any effect in adult kidney, heart and liver (16). The group hypothesised that THOC5 was essential for the maintenance of stem cells/progenitor cells but not for the terminally differentiated cells like hepatocytes. The same group demonstrated elevated levels of THOC5 in HCC tissues and cell lines, alongside an induction of apoptosis and cell cycle deregulation in vitro, after THOC5 knock down (17). Hypothesising that THOC5 target genes were important in liver cancer cell survival, Tamura’s team went on to study one target gene in particular—namely the lncRNA 00176 (Linc00176).
Linc00176 (NR_027686.1) is located on chromosome 20 and its HCC transcript is reported to have 4 exons and be 5,264 nucleotides long. In the HepG2 HCC cell line an alternatively spliced lnc00176 transcript, lacking exon 1, 1,601 nucleotides from exon 2 as well as 962 nucleotides from exon 4, has been identified (18). Apart from this information, little was known about the role of Linc00176 in HCC. Tran et al. (10) have now explored HCC patients data available from The Cancer Genome Atlas (TCGA) and the ENCODE consortium (18). Linc00176 alternatively spliced transcripts were present in HepG2, Hep3B, Huh7, HLE and HLF HCC cell lines, with HepG2 cells showing the highest expression levels. Linc00176 expression wasn’t evident in any normal human tissues (including liver, pancreas, lung, skin, brain, adipose tissue, muscle, heart and bone marrow) or other malignancies (leukaemia, melanoma, breast cancer, neuroblastoma, rhabdomyosarcoma and cervical human cancer cell lines) (10). In human primary HCC data in TCGA, high versus low levels of Linc00176 expression was significantly associated with both poorly differentiated tumour grade and poorer patient survival.
Studying Linc00176 putative promoter region (500 nucleotides upstream of the initiation site), Tran et al. have integrated human hepatocyte and HepG2 cells DNase-Seq, RNA-Seq, ChIP seq and cap analysis of gene expression (CAGE) data (18) and applied PROMO (ALGGEN, program predicting transcription factor binding sites) software. The team focused on a number of candidate regulatory transcription factors (Myc, MAZ and AP-4). Myc was reported to preferentially regulate the transcription machinery of lncRNAs (19) and Tran et al. have expanded this observation in vitro by showing that Myc/AP-4 double knock down synergistically depleted Linc00176 expression (10). Functionally, depletion of Linc00176 from Huh7 and HepG2 abolished their proliferation and converted them to TUNEL-positive cells after 2 days compared to control cells. Unlike THOC5 depletion-induced apoptosis (17), Linc00176 reportedly to exerts its role through necroptosis via mixed lineage kinase-like (MLKL)(10). The bioinformatics ingenuity pathway analysis (IPA) performed on the genes differentially expressed in HepG2 cells after knock down of Linc00176 supported this finding, ranking cancer and necrosis as the top deregulated pathways.
Having identified upstream regulators of Linc00176, as well as the impact of its knockdown, Tran et al. considered the three main mechanisms by which lncRNAs bring about their regulatory functions (20). Some lncRNAs act as physical scaffolds forming flexible macromolecular complex with nuclear matrix, chromatin regulatory and DNA methylation proteins to control the chromatin state. Other lncRNAs recruit chromatin regulatory machinery to specific DNA loci either through their affinity to some regulatory proteins or through 3D proximity-guided localisation to certain gene loci. Alternatively, many lncRNAs shape the nuclear structure in a way that encourage/discourage gene expression. Tran et al. went on to demonstrate that Linc00176 binds to two anti-tumour, Myc-regulated miRNAs, namely miRNA-9 and miRNA-185 (10). Depleting the concentrations of these miRNAs diminished their anti-proliferative effects on the tumour cells. Moreover, depletion of these two miRNAs using specific inhibitors rescued the proliferation-prohibited effect of Linc00176 knock down in tumour cell lines.
This work by Tamuras’ group (10) exploits publicly available datasets, combined with confirmatory and exploratory laboratory studies, in a fashion that brings lncRNAs to the heart of the HCC field. Not only are these transcripts highly specific for HCC, suggesting potential roles in HCC specific diagnosis, their functional roles also raise the potential for intervention to either prevent or treat cancers. Linc00176 joins other lncRNAs, like previously reported RP11-166D19.1 (13), as a candidate diagnostic biomarker. In addition, this study suggests that targeting of Linc00176 would not only selectively inhibit the proliferation of tumour cells, it would also favour the development of an anti-tumour niche by increasing the availability of tumour-limiting miRNAs like miRNA-9 and miRNA-185. The development of diagnostic and monitoring biomarker assays, measuring Linc00176 and its targets in tissues, alongside the means to target its functional effects, may be worthy of consideration for patients with HCC. There may be other lncRNAs, or combinations, which have relevance to different patients, possibly with cancers arising in different etiological backgrounds. This is a landmark study not just because it highlights the potential importance of Linc00176, but because it leads the way in HCC, demonstrating the translational value of systematic interrogation and integration of publicly available data (Figure 1).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Meiyi Song (Division of Gastroenterology and Hepatology, Digestive Disease Institute, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.03.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Reeves HL, Zaki MY, Day CP. Hepatocellular Carcinoma in Obesity, Type 2 Diabetes, and NAFLD. Dig Dis Sci 2016;61:1234-45. [Crossref] [PubMed]
- Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol 2010;7:448-58. [Crossref] [PubMed]
- Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;389:56-66. [Crossref] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009;69:7385-92. [Crossref] [PubMed]
- Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 2008;359:1995-2004. [Crossref] [PubMed]
- Llovet JM, Villanueva A, Lachenmayer A, et al. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015;12:436. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network, Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref] [PubMed]
- Tran DD, Kessler C, Niehus SE, et al. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene 2018;37:75-85. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208. [Crossref] [PubMed]
- Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun 2017;8:14421. [Crossref] [PubMed]
- Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012;22:1760-74. [Crossref] [PubMed]
- Katahira J. mRNA export and the TREX complex. Biochim Biophys Acta 2012;1819:507-13.
- Mancini A, Niemann-Seyde SC, Pankow R, et al. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol 2010;8:1. [Crossref] [PubMed]
- Saran S, Tran DD, Ewald F, et al. Depletion of three combined THOC5 mRNA export protein target genes synergistically induces human hepatocellular carcinoma cell death. Oncogene 2016;35:3872-9. [Crossref] [PubMed]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799-816. [Crossref] [PubMed]
- Zheng GX, Do BT, Webster DE, et al. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol 2014;21:585-90. [Crossref] [PubMed]
- Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756-70. [Crossref] [PubMed]
Cite this article as: Zaki MYW, Reeves HL. Long intergenic non-coding RNAs in hepatocellular carcinoma—a focus on Linc00176. Non-coding RNA Investig 2018;2:12.


