
Inhibition of miR-223 reduces inflammation but not adverse cardiac remodelling after myocardial ischemia-reperfusion in vivo
Introduction
Acute myocardial infarction (MI) remains one of the leading causes of morbidity and mortality in the western world (1).Coronary artery occlusion results in ischemic heart tissue and subsequent death of cardiomyocytes, followed by an inflammatory response to clear the infarct from dead cells (2,3). Currently, the most effective therapy to salvage dying myocytes is early reperfusion, although reperfusion itself also leads to additional damage known as myocardial ischemia-reperfusion (I/R) injury (4). Although the exact mechanism still remains to be established, generation of reactive oxygen species (ROS) by local endogenous cells and a rapid influx of inflammatory cells and cytokines seems to play an important role (5). Consequently, the role of inflammatory cells and its cytokine production in infarct healing has been investigated thoroughly during the last decades as extensively reviewed elsewhere (6). Unfortunately, therapeutic interventions did not lead to desirable results, mostly due to the complex spatial and temporal role of inflammation during the process of myocardial wound healing (7). Given the importance of the inflammatory phase during cardiac wound healing, better understanding as well as novel interventions still needs to be explored.
MicroRNAs (miRNAs) are short, non-coding small RNAs that regulate the expression of proteins by translational repression or degradation of messenger RNAs (mRNAs) (8). Over the last few years it has become clear that miRNAs play a crucial role in cardiac development and homeostasis and that miRNA expression is altered in the diseased heart (9-11). Not surprisingly, miRNAs are increasingly seen as master regulators of many processes, including immune cell function and inflammation (12).
To examine the role of miRNAs during wound healing in the heart, we profiled the expression of several miRNAs after I/R injury. One interesting candidate was miR-223. Interestingly, this miRNA is considered to play a role in insulin metabolism (13), coronary artery disease (14), hypercholesterolemia (15), and importantly, in the regulation of neutrophil and macrophage response to inflammatory stimuli (16).
In the present study, we demonstrate that miR-223 is differentially expressed during the inflammatory phase after cardiac ischemia. Furthermore, we determined the effect of miR-223 inhibition on inflammation and cardiac remodelling in a mouse model of myocardial I/R.
Methods
This study was approved by the Animal Ethical Experimentation Committee (Utrecht University) and was carried out in accordance with the Guide for the Care and Use of Laboratory Animals (17).
Murine model of myocardial I/R
Male C57Bl/6 mice (10–12 weeks, Harlan Laboratories) underwent left coronary artery (LCA) ligation as previously described (18), followed by reperfusion. Briefly, mice were anesthetized with fentanyl (0.05 mg/kg), midazolam (5 mg/kg), and medetomidine (0.5 mg/kg) through an intraperitoneal (ip) injection, and the adequacy of anaesthesia was monitored by disappearance of the pedal withdrawal reflex. Mice were ventilated with 100% oxygen, maintaining a core body temperature of 37 °C. The LCA was ligated just below the left atrial appendage for 30 minutes with an 8-0 Ethilon monifil suture, including a piece of soft tubing. Reperfusion was initiated by releasing the ligature and removal of the tubing, leaving a piece of suture in place. The chest wall was closed in layers and the animals received atipamezole (2.5 mg/kg ip), flumazenil (0.5 mg/kg ip), and temgesic (0.1 mg/kg ip) for pain killing. After detubation, mice were kept warm until fully recovered. At different times, mice were sacrificed using a cocktail of ketamine (100 mg/kg ip) and medetomidine (8 mg/mL), followed by cardiac explantation.
Micro-array
After myocardial I/R, total RNA was isolated with the mirVana RNA Isolation Kit (Ambion). MiRNA expression profile was determined by microarray using the Paraflo microfluidic chip (MiHuman_8.2, LC Sciences), according to the manufacturer, and validated with Taqman MicroRNA Assays (Applied Biosystems).
Quantitative RT-PCR of miRNAs
Total RNA was isolated from cells, total left ventricle (LV), or other organs using TriPure Isolation Reagent (Roche Applied Science, Penzberg, Germany). Ten nanogram DNA-free RNA was used for miRNA-specific (miR-223; assay ID 002295, Applied Biosystems) reverse transcription and subsequent amplification and detection in a MyIQ single-colour quantitative real time polymerase chain reaction (qRT-PCR) system (Bio-Rad, Hercules, CA) as described previously (19). After normalization against the small nuclear RNA RNU19 (assay ID 001003, Applied Biosystems), the relative difference in expression levels was calculated and presented as fold induction (2ΔΔCt).
Isolation of neonatal mouse cardiomyocytes
Neonatal mouse cardiomyocytes were isolated and cultured as reported previously (20). Briefly, after termination by decapitation, five 1–3 days old mouse hearts were cut into small pieces and digested by 0.5% trypsin to form a single cell suspension. This suspension was incubated another 5 minutes in a stirring water bath at 37 °C, which was repeated three times after washing in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Carlsbad), supplemented with 20% fetal calf serum (FCS) (Gibco). After resuspending the cell pellet in culture medium of DMEM with 20% FCS, 5% horse serum (Gibco), penicillin (100 U/mL), and streptomycin (100 mg/mL), cells were pre-plated in an uncoated dish for 2.5 hours, replated in 1% gelatine at a density of 1.5×105 cells/cm2 and left to attach over night at 37 °C before any experiment was started.
Cardiomyocyte viability assay
Neonatal mouse cardiomyocytes were pre-incubated with culture medium (DMEM with penicillin and streptomycin) without serum for one hour and challenged with 50 µM H2O2 or DMSO (control) for 18 hours, after which the cells were washed and dissolved in TriPure Isolation Reagent (Roche Applied Science, Penzberg, Germany) and stored at −80 °C.
Antagomirs
Antagomirs were designed as previously described and custom synthesized (VBC Biotech, Vienna, Austria) (21). Antagomir sequences used were: antagomir-223 (5`- ugggguauuugacaaacugaca -3`), and antagomir-mismatch-ctrl (5`- uggcguaucugucauacagaaa -3`). The mismatch control antagomir (ctrl-antagomir) contained 6 mismatches, all antagomirs were 3`cholesterol-modificated, 2`O-methylated, and contained PTO-linkages at the first two and last four nucleotides. The mismatch control antagomir did not target any other known miRNAs.
Antagomir treatment
Mice underwent intravenous (tail vein) injections of 8 mg/kg body weight antagomir 5 min prior to reperfusion and 1 and 2 days later. The efficiency of miRNA knockdown was confirmed by TaqMan MicroRNA Assay either 3 (short term follow-up) or 28 days (long term follow-up) after initial injection.
Magnetic resonance imaging (MRI)
Mice underwent serial assessment of cardiac dimensions and function by MRI (9.4T MRI; Bruker, Germany) under isoflurane anaesthesia before and 28 days after surgery (n=7/group) as described previously (22). LV function and geometry was assessed by a blinded observer. All MRI data were analyzed with Qmass digital imaging software (Medis, Leiden, the Netherlands).
In situ hybridization
In situ hybridization was performed to determine hsa-miR-223 localization as described previously (19). Briefly, 10 µm thick cryosections were hybridized O/N at 46 °C with 5 nM LNA DIG-labelled probes (Scramble-miR, Exiqon, 99001-01 and hsa-miR-499, Exiqon, 38500-01) in denaturizing hybridization buffer, incubated with an anti-DIG-alkaline phosphatase antibody (1:1,500, Roche 11093274910), an anti-troponin antibody (Sigma) at RT for 2 hours. After washing, sections were incubated with fluorescent-labelled secondary antibodies, exposed to Fast-Red Substrate-Chromogen (DAKO, K0597) O/N at RT and mounted in VectaShield (Molecular Probes, Amsterdam).
Immunohistochemistry and histology
After termination at 3 days post-IR, hearts were flushed with 0.9% saline, explanted and snap-frozen in liquid nitrogen. Frozen heart sections of 7 µm were stained for neutrophils (rat anti-mouse Ly6G, BD Pharmingen 551459) and macrophages (rat anti-mouse Mac-3, BD Pharmingen 553322). Secondary antibody used was goat anti-rat IgG (Southern Biotech, 3052-08) followed by HRPO-coupled streptavidin (Southern Biotech, 7100-05), using 3-Amino-9-ethyl carbazole (AEC) for visualisation.
Statistical analysis
Data are presented as mean ± SEM. Differences between groups were analyzed using ANOVA with Bonferroni correction or Mann-Whitney U test when appropriate. All tests were two-sided, using a significance level of P<0.05 (SPSS Statistics v17Chicago, USA).
Results
MiR-223 expression is increased after I/R in vivo
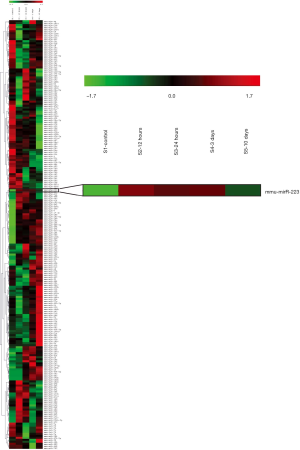
To examine the role of miRNAs during wound healing in the heart, we performed a micro-array on several miRNAs after cardiac I/R injury (Figure S1). Interestingly, miR-223 showed increased levels after 1 and 3 days post I/R compared to baseline (Figure 1A). At 10 days, miR-223 levels were almost returned to basal levels. Micro-array data on miR-223 was validated using RT-PCR, showing a significant increase after 1 and 3 days, corresponding with the inflammatory phase after cardiac injury (3) (Figure 1B).
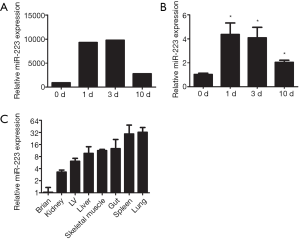
Furthermore, we determined the tissue expression of miR-223 in the heart compared to other mouse organs. MiR-223 expression was most obvious in spleen and lung tissue, which contain large numbers of monocytes and alveolar macrophages, respectively (Figure 1C). MiR-223 levels were the lowest in brain tissue and moderately expressed in skeletal muscle and liver.
Mir-223 is highly expressed in cardiomyocytes after I/R in vivo
We performed in situ hybridization to determine in which cells miR-223 was localized. At baseline, only a small number of cells showed peri-nuclear staining for miR-223 (Figure 2A), which was comparable with the remote area three days post-I/R (Figure 2B). However, miR-223 staining was markedly increased in the border zone (Figure 2C) in cardiomyocytes predominantly and to a lesser extent in non-myocytes. In situ hybridization of the negative control (scramble-miR) did not result in any staining of the border zone (Figure 2D).
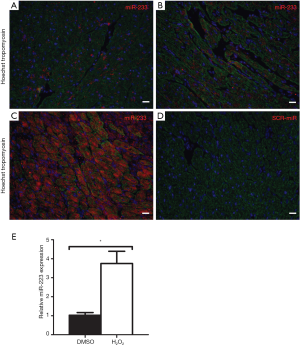
To verify whether the increased expression of miR-223 could result from stressed cardiomyocytes rather than from invading inflammatory cells, freshly isolated neonatal mouse cardiomyocytes were challenged by H2O2 stimulation to assess the effect of oxidative stress. Compared to normal conditions, oxidative stress resulted in a 3.75-fold increase of miR-223 expression, as quantified by RT-PCR (Figure 2E, P<0.05).
Inhibition of miR-223 reduced inflammatory cell influx after myocardial I/R in vivo
Considering the increased miR-223 expression after I/R, we investigated the effect of its modulation in vivo using an antagomir or a mismatch (scramble-miR) control. As a first step, we determined miR-223 levels, by RT-PCR after a 3-day inhibition protocol in several organs, including the heart. Mice treated with antagomir-223 displayed a significant reduction of up to 99% in miR-223 expression after 3 days of the initial injection (Figure 3A).
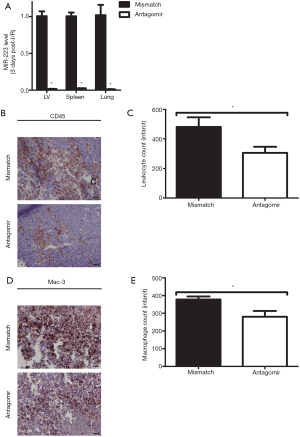
As our previous results showed that miR-223 mainly increased during the inflammatory phase after I/R, we studied whether its inhibition influenced the influx of inflammatory cells. Antagomir–treated animals showed a reduction of leukocyte influx compared to mismatch treatment as quantified by CD45 staining (Figure 3B,C; P<0.05). In accordance with a reduction of total inflammatory influx, antagomir treatment significantly reduced macrophage influx at 3 days of reperfusion as well (Figure 3D,E).
Inhibition of miR-223 does not influence adverse remodelling after myocardial I/R in vivo
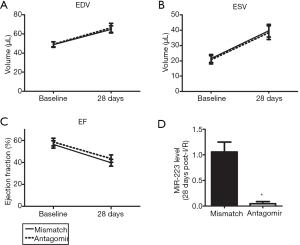
Invading leukocytes contribute to myocardial I/R injury and adverse remodelling (4,5). Since inhibition of miR-223 inhibited the inflammatory influx into the myocardium, we assessed the effect of antagomir treatment on cardiac function and remodelling. End-diastolic volume (EDV) and end-systolic volume (ESV) of both treatment groups increased significantly after myocardial I/R at 28 days (Figure 4A,B). Antagomir administration did not preserve LV volumes compared to mismatch treatment. As a consequence, the decline in ejection fraction (EF) post-I/R was similar in both the treatment groups (Figure 4C). PCR revealed that miR-223 knockdown was still achieved at 28 days after the Antagomir injections (Figure 4D).
Discussion
In this study, we have demonstrated that miR-223 increased after myocardial I/R in vivo, particularly in cardiomyocytes during day 1 and 3, which was confirmed by in situ hybridization and mimicked in in vitro experiments. Furthermore, we were able to inhibit miR-223 using an antagomir resulting in an ~99% knockdown in vivo. Inhibition of miR-223 reduced the influx of inflammatory cells and macrophages in the myocardium after I/R, but did not lead to a long term functional benefit in terms of cardiac function.
Initial reports on miR-223 function were mostly related to a role in granulocyte and monocyte maturation and differentiation (23,24). Yu et al. were one of the first suggesting a role for miR-223 in ischemic disease, reporting an increase if miR-223 expression after hepatic I/R injury (25), which was followed by reports showing increased miR-223 expression in Duchenne muscle dystrophy and spinal cord injury, including co-expression on invading neutrophils (26-28). Cardiac miR-223 levels were previously shown to be increased in LV biopsies of patients with type 2 diabetes, positively regulating Glut4 expression on cardiomyocytes (13).
In line with a previous report investigating miRNA changes in a mouse model of permanent coronary artery occlusion (29), we report increased levels of miR-223 after cardiac I/R. Interestingly, this was most obvious in the cardiomyocytes themselves. During MI and cardiac ischemia, it was suggested that increased glucose uptake could lead to the activation of Akt leading to BAD-phosphorylation and subsequently to an increase in cell survival (30). As miR-223 was suggested to increase GLUT4 expression (13), increased myocardial miR-223 levels might lead to increased glucose uptake, thereby promoting cell survival.
It was shown previously that neutrophils lacking miR-223 have an increased oxidative burst making them more effective against hostile pathogens, showing that miR-223 negatively regulates their activation (23). Very recently, it was reported that mice with a total miR-223 knock-out developed a more pro-inflammatory macrophage setup, increasing M1 subtype macrophages and leading to an increased inflammatory response (31). Although we used a model of cardiac ischemia, our results demonstrate that miR-223 inhibition leads to decreased levels of leukocytes and especially macrophages. Although we did not further characterize the subtype of macrophages, miR-223 modulation neither improved nor lead to a further decline in long-term cardiac performance compared to control animals. One explanation could be that the inhibitory effect was long-lasting where inhibition in the early inflammatory phase only could be beneficial.
It remains to be seen whether miR-223 modulation can lead to functional improvements after cardiac I/R as several questions need to be addressed first. As with previous attempts to modulate the process of inflammation after myocardial I/R, close attention must be paid to the spatial and temporal role of miR-223 during the process of myocardial wound healing. It could well be that the effect of miR-223 modulation on cardiomyocytes (i.e., inhibition leading to increased survival) is opposite to the effect on inflammatory cells (i.e., inhibition leading to a more pro-inflammatory subset). Although preliminary, the results from the above study suggest that general miR-223 inhibition after I/R, involving many cell types in a period of time that exceeds the inflammatory phase, will not lead to a net beneficial effect in the ischemic heart. More in depth studies on the complex temporal and spatial role of miR-223 during the process of myocardial wound healing seems necessary in order to translate this interesting concept into a novel therapeutic strategy.
Acknowledgments
We thank the following persons for their excellent assistance: Corina Metz, and Arjan Schoneveld (all at the University Medical Center Utrecht, Utrecht, the Netherlands).
Funding: This work was supported by the Novartis Foundation for Cardiovascular Excellence (JS), a Bekalis price (PD), and “Stichting Swaeneborgh” (MO).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.03.04). This work was published before but that the journal was discontinued. JPS serves as an unpaid editorial board member of Non-coding RNA Investigation from February 2018 to January 2020. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Animal Ethical Experimentation Committee (Utrecht University) (approval ID: DEC2010.II.04.086) and was carried out in accordance with the
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Writing group members. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation 2010;121:e46-e215. [Crossref] [PubMed]
- Frantz S, Bauersachs J, Ertl G. Post-infarct remodelling: contribution of wound healing and inflammation. Cardiovasc Res 2009;81:474-81. [Crossref] [PubMed]
- Cleutjens JP, Blankesteijn WM, Daemen MJ, et al. The infarcted myocardium: simply dead tissue, or a lively target for therapeutic interventions. Cardiovasc Res 1999;44:232-41. [Crossref] [PubMed]
- Oerlemans MI, Koudstaal S, Chamuleau SA, et al. Targeting cell death in the reperfused heart: pharmacological approaches for cardioprotection. Int J Cardiol 2013;165:410-22. [Crossref] [PubMed]
- Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N Engl J Med 2007;357:1121-35. [Crossref] [PubMed]
- Frangogiannis NG. Regulation of the inflammatory response in cardiac repair. Circ Res 2012;110:159-73. [Crossref] [PubMed]
- Frangogiannis NG. Targeting the inflammatory response in healing myocardial infarcts. Curr Med Chem 2006;13:1877-93. [Crossref] [PubMed]
- Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell 2003;115:787-98. [Crossref] [PubMed]
- Sluijter JP, van Mil A, van Vliet P, et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol 2010;30:859-68. [Crossref] [PubMed]
- Thum T, Galuppo P, Wolf C, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 2007;116:258-67. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255-60. [Crossref] [PubMed]
- O'Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood 2011;118:2960-9. [Crossref] [PubMed]
- Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res 2010;86:410-20. [Crossref] [PubMed]
- Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810-7. [Crossref] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Schroen B, Heymans S. Small but smart--microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res 2012;93:605-13. [Crossref] [PubMed]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. 8th edition. Washington (DC): National Academies Press (US), 2011.
- Oerlemans MI, Goumans MJ, van Middelaar B, et al. Active Wnt signaling in response to cardiac injury. Basic Res Cardiol 2010;105:631-41. [Crossref] [PubMed]
- van Mil A, Grundmann S, Goumans MJ, et al. MicroRNA-214 inhibits angiogenesis by targeting Quaking and reducing angiogenic growth factor release. Cardiovasc Res 2012;93:655-65. [Crossref] [PubMed]
- Sreejit P, Kumar S, Verma RS. An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell Dev Biol Anim 2008;44:45-50. [Crossref] [PubMed]
- Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 2005;438:685-9. [Crossref] [PubMed]
- Noort WA, Oerlemans MI, Rozemuller H, et al. Human versus porcine mesenchymal stromal cells: phenotype, differentiation potential, immunomodulation and cardiac improvement after transplantation. J Cell Mol Med 2012;16:1827-39. [Crossref] [PubMed]
- Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008;451:1125-9. [Crossref] [PubMed]
- Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 2005;123:819-31. [Crossref] [PubMed]
- Yu CH, Xu CF, Li YM. Association of MicroRNA-223 expression with hepatic ischemia/reperfusion injury in mice. Dig Dis Sci 2009;54:2362-6. [Crossref] [PubMed]
- Greco S, De Simone M, Colussi C, et al. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J 2009;23:3335-46. [Crossref] [PubMed]
- Nakanishi K, Nakasa T, Tanaka N, et al. Responses of microRNAs 124a and 223 following spinal cord injury in mice. Spinal Cord 2010;48:192-6. [Crossref] [PubMed]
- Izumi B, Nakasa T, Tanaka N, et al. MicroRNA-223 expression in neutrophils in the early phase of secondary damage after spinal cord injury. Neurosci Lett 2011;492:114-8. [Crossref] [PubMed]
- van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027-32. [Crossref] [PubMed]
- Kalra BS, Roy V. Efficacy of metabolic modulators in ischemic heart disease: an overview. J Clin Pharmacol 2012;52:292-305. [Crossref] [PubMed]
- Zhuang G, Meng C, Guo X, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 2012;125:2892-903. [Crossref] [PubMed]
Cite this article as: Oerlemans MIFJ, van Mil A, Liu J, van Eeuwijk E, den Ouden K, Doevendans PA, Sluijter JPG. Inhibition of miR-223 reduces inflammation but not adverse cardiac remodelling after myocardial ischemia-reperfusion in vivo. Non-coding RNA Investig 2018;2:15.


