
Long non-coding RNAs are driver to maintain the chromatin active regions at divergent transcriptional units
Understanding of the diverse role of the human long non-coding RNAs (lncRNAs) is a considerable emerging field in development and disease (1). LncRNAs are broadly defined as noncoding RNAs longer than 200 nucleotides, separating them from various classes of microRNAs (miRNAs), small nuclear RNAs (snRANs), and small nucleolar RNAs (snoRNAs) that function through distinct mechanisms (2,3). The majority of lncRNAs are transcribed by RNA polymerase II (RNAP2), hence sharing similarities with messenger RNAs (mRNAs). However, lncRNAs are characterized by lack of coding potential, week co-transcriptional splicing, and are mostly restricted to chromatin (4,5). To date, the ENCODE project (GENCODE v26) analysis have identified approximately 16,000 human lncRNA genes that correspond to more than 28,000 distinct transcripts in the cell (6). However, how many of them are functional is still unclear. Previous work has revealed that lncRNAs fulfil multiple key regulatory roles in gene expression from guiding epigenetic modifications to modulating mRNA stability and translation (7-9). Therefore, lncRNAs are being considered as a major class of regulatory elements in the genome.
Recent evidence suggested that lncRNAs could function through binding to histone-modifying complexes, transcription factors, and also the RNA polymerase II (10,11). Indeed, lncRNAs act as guides and sponges for chromatin regulators, most likely titrating RNA and proteins and hence contribute to fine tuning the chromatin organization in the genome (12). There is evidence supporting the ability of lncRNAs to collaborate with chromatin modifiers in a spatio-temporal fashion within the distinct compartments (13-15). The lncRNA HOTTIP binds the adaptor protein WDR5 (WD repeat-containing protein 5) and targets WDR5/MLL complexes to maintain the histone H3 lysine 4 trimethylation (H3K4me3), and coordinate homeotic gene expression. On the other hand, another lncRNAs binds EZH2, the catalytic subunit of the PRC2 complex, and decreased the H3K27me3 mark (16). These observations suggest that different lncRNAs exhibit special features in their spatio-temporal conformation, localization or length, which allow them to sense the transcriptional state of the cell and to target the chromatin modifiers or remodelers across the genome.
Consistent with this notion, the article of Subhash et al. in a recent issue of Nuclei Acids Research (17) offers the example of an active Chromatin-Associated lncRNAs (lncCARs) isolated using chromatin RNA immunoprecipitation followed by deep sequencing (ChRIP-eq) (18) from chromatin enriched in H3K4me2 and WDR5. The authors identified the lncCARS in the breast cancer cell line BT-549 treated with Actinomycin D to avoid co-transcriptional crosslinking of lncRNAs and chromatin. Initially, they identified 544 H3K4me2-lncRNAs (enriched by two-fold over the nuclear input) and found then approximately 70% of them were present in an antisense orientation (designated “X”) to their partner protein-coding gene. Moreover, the majority of antisense transcripts coincides with divergent orientation compared to non-CARs lncRNAs. Overall, they conclude that H3K4me2 lncRNAs are predominantly organized as antisense diversion (H3K4me2-XH-lncRNAs) units and regulate the expression of their adjacent partner protein-coding genes.
Given the fact that H3K4me2 bind directly with WDR5 (known to catalyze the conversion of H3K4me2 to H3K4me3) a critical component of chromatin remodeling complex such as MLL/SET1 and also shown to bind lncRNAs to coordinate homeotic gene expression (15,19). Therefore, Subhash and colleagues investigated WDR5-enriched chromatin interacting RNAs in BT-549 cells using the ChRIP-seq and found 807 WDR5-lncRNAs. Similar to H3K4me2-lncRNA, they found that approximately, 70% of WDR5-lncRNAs organized in antisense orientation to their partner protein coding gene. They found 209 of H3K4me3-lncRNAs shared by WDR5-lncRNAs and also carrying specific feature of antisense divergent transcription unit (H3K4me3/WDR5-XH-lnCARs), so they code them as active XH transcription units. Also, divergent transcription organization feature was underrepresented in case of inactive lncCARs in BT-549 cells define by H3K27me3/EZH2 enrichment, highlighting specific organization of active XH transcription units in the genome.
In a search for the functional significance of active XH transcriptional units, they observed more enrichment of H3K4 methylation (me2 and me3) as well WDR5 at the promoter region of active lncCARs in comparison to non-chromatin associated RNAs (CARs) with divergent (XH) pattern (non-CARs XH-lncRNAs). Furthermore, they showed that active XH-lncCARs interacts to the WDR5 and this interaction is compromised in the presence of a F266A WDR5 mutant that affect the RNA binding motif of WDR5. The question arises whether active XH lncCARs contribute to maintain the transcriptional competence at the adjacent partner protein coding genes promoter? To explore this possibility, the authors performed chromatin oligo affinity precipitation (ChOP) assay (20) and confirmed that active XH lncCARs is recruited to their respective protein coding partner promoters to maintain their expression. Next, they systematically showed that downregulation of active XH lncCARs leads to a significant decrease in the levels of the H3K4me2, H3K4me3 marks as well as in the WDR5 binding to their respective paired protein coding promoter regions. Finally, the authors demonstrated that WDR5 is required for the conversion of H3K4me2 to H3K4me3 specifically at the promoter region of active transcription units.
In summary, Subhash et al. characterized a new category of active (H3K4me3/WDR5) XH-lncCARs and functionally demonstrated that the presence of XH lncCARs is required to maintain the transcriptionally competent chromatin at their paired protein coding genes and that WDR5 establishes the H3K4me3 mark at XH-lncCARs promoters. Notably, authors showed that transient overexpression of target XH-lncCAR cannot activate the expression of their divergent protein coding partner gene, suggesting that XH-lncCAR needs to be transcribed in cis to fulfil its functional effect on protein coding genes. Given the fact that WDR5 is predicted to contain intrinsically disordered regions (IDRs) that are conserved through vertebrate species (21) these findings open the possibility that WDR5 alone or in combination with of other low complex domain (LCD) proteins of the transcription machinery favors the formation liquid-liquid phase transition (LLPS) with RNAP2-CTD (22,23) and proposed model presented in Figure 1.
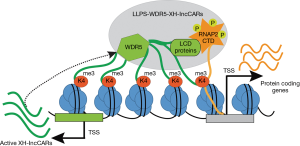
Future studies will require to prove this hypothetical model either by testing XH-lncCARs directly binding to RNA binding motif of WDR5 or by identifying its interaction with other LCD proteins that eventually guide their recruitment to the LLPS encompassing the active transcription machinery, which will act on their divergent protein coding partner genes. A comprehensive analysis of distinct sets of lncRNAs and their associated protein coding genes will be required to define the generality of this gene regulation mechanism.
Acknowledgments
Funding: The authors supported by Spanish MEC (SAF2013-42497), the Catalan Government (AGAUR 2014SGR780) and the European Research Council Synergy Grant “4DGenome” (609989). We also acknowledge the support of the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa” and the CERCA Programme/Generalitat de Catalunya”.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Jin Li (Cardiac Regeneration and Ageing Lab, School of Life Sciences, Shanghai University, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.08.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. [Crossref] [PubMed]
- Uszczynska-Ratajczak B, Lagarde J, Frankish A, et al. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208. [Crossref] [PubMed]
- Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 2018;19:143-57. [Crossref] [PubMed]
- Schlackow M, Nojima T, Gomes T, et al. Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol Cell 2017;65:25-38. [Crossref] [PubMed]
- Hon CC, Ramilowski JA, Harshbarger J, et al. An atlas of human long non-coding RNAs with accurate 5' ends. Nature 2017;543:199-204. [Crossref] [PubMed]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300-7. [Crossref] [PubMed]
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [Crossref] [PubMed]
- de Andres-Pablo A, Morillon A, Wery M. LncRNAs, lost in translation or licence to regulate? Curr Genet 2017;63:29-33. [Crossref] [PubMed]
- Long Y, Wang X, Youmans DT, et al. How do lncRNAs regulate transcription? Sci Adv 2017;3:eaao2110.
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Chen CK, Blanco M, Jackson C, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 2016;354:468-72. [Crossref] [PubMed]
- Zhao J, Ohsumi TK, Kung JT, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010;40:939-53. [Crossref] [PubMed]
- Gomez JA, Wapinski OL, Yang YW, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 2013;152:743-54. [Crossref] [PubMed]
- Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011;472:120-4. [Crossref] [PubMed]
- Kaneko S, Son J, Shen SS, et al. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 2013;20:1258-64. [Crossref] [PubMed]
- Subhash S, Mishra K, Akhade VS, et al. H3K4me2 and WDR5 enriched chromatin interacting long non-coding RNAs maintain transcriptionally competent chromatin at divergent transcriptional units. Nucleic Acids Res 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Mondal T, Subhash S, Kanduri C. Chromatin RNA Immunoprecipitation (ChRIP). Methods Mol Biol 2018;1689:65-76. [Crossref] [PubMed]
- Wysocka J, Swigut T, Milne TA, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005;121:859-72. [Crossref] [PubMed]
- Akhade VS, Arun G, Donakonda S, et al. Genome wide chromatin occupancy of mrhl RNA and its role in gene regulation in mouse spermatogonial cells. RNA Biol 2014;11:1262-79. [Crossref] [PubMed]
- Lazar T, Schad E, Szabo B, et al. Intrinsic protein disorder in histone lysine methylation. Biol Direct 2016;11:30. [Crossref] [PubMed]
- Lu H, Yu D, Hansen AS, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018;558:318-23. [Crossref] [PubMed]
- Boehning M, Dugast-Darzacq C, Rankovic M, et al. RNA polymerase II clustering through CTD phase separation. bioRvix 2018. doi: https://doi.org/
10.1101/316372
Cite this article as: Sharma P, Beato M. Long non-coding RNAs are driver to maintain the chromatin active regions at divergent transcriptional units. Non-coding RNA Investig 2018;2:50.


