
LncRNA miR503HG is a new player in hepatocellular carcinoma metastasis
Recently, lncRNA have been found to be involved in many biological processes (1-4). They have been discovered in all of the hallmarks of cancer (5,6) and many are used as biomarkers and therapeutic targets in specific cancers. Therefore, it is not surprising that their dysregulation has been found to lead to disease and progression of diseases such as cancer (6-9). As the third-leading cause of cancer death worldwide, it is important to study the underlying mechanisms related to hepatocellular carcinoma (HCC) progression (10). Though surgery is an effective treatment option for HCC, many patients succumb to the disease due to recurrence and metastasis (11,12). This is why prognostic biomarkers are so important to the treatment of cancer. They give doctors a way to determine and select the best path of treatment and what can provide (13,14).
Recently Wang et al. reported that a lncRNA known as miR503HG is responsible for inhibiting metastasis in HCC through its regulation of the heterogeneous nuclear protein (hnRNP) hnRNPA2B1 mediated NF-κB pathway (15). In their assessment of a large group of HCC patient samples compared to paracancer controls, 713 lncRNA were found to be dysregulated. The authors then narrowed the search to differentially regulated lncRNA that were transcribed from host genes to microRNA and found five lncRNA that fit this criterion. Only one of these lncRNA were found to be significantly dysregulated (downregulated) in HCC, miR503HG. To support the importance of this lncRNA in HCC, a Kaplan-Meier analysis of patients was performed and indicated that miR503HG is an important factor for survival after surgery as well as decreasing the probability of recurrence after surgery. In order to understand the relationship between patient prognosis and miR503HG, Wang et al. selected two HCC cell lines for migration and metastasis experiments in vitro and in vivo. The two cell lines are known as SMMC-7721 and Huh7 and have decreased expression of miR503HG lncRNA, but still much lower than a normal liver cell line. These cell lines allowed for the knock down of miR503HG, which exacerbated migration and invasion severity indicating miR503HG plays a role in HCC metastasis. Overexpression of the lncRNA led to a drastic decrease in both in vitro and in vivo migration and invasion demonstrating that miR503HG is involved in the suppression of metastasis. Also of note, it was discovered that the microRNA, miR503, which is also expressed from the host gene of miR503HG, can work synergistically with miR503HG to mediate the suppression of cell migration and invasion. However, the two noncoding RNAs use independent pathways to suppress metastasis. This microRNA has been studied in HCC previously and it has been implicated as a player in the inhibition of proliferation and induction of apoptosis in HCC (16). This likely means that miR503 could be involved in multiple pathways to aid in the regulation of cancer progression. Since miR503HG is also transcribed from the same gene, it may also play a role in other cancer suppression pathways as well. In total, these experiments revealed the important overall biological function of miR503HG to be suppressing metastasis in HCC.
After uncovering the overall functional importance of miR503HG, Wang et al. investigated the mechanism through which miR503HG acts on metastasis in HCC. First, the group wanted to identify any direct protein interactions with the lncRNA. In an RNA pull-down assay using HCC cell extracts and mass spectrometry, it was discovered that 25 proteins potentially interacted with miR503HG. It has been shown previously that lncRNA can interact with hnRNPs in signaling pathways and to effect chromatin regulation (17,18). Therefore, the hnRNP, hnRNPA2B1, was selected from among the 25 interacting proteins and further investigated. This hnRNP was confirmed as a specific binding partner for miR503HG through western blot analysis and RNA immunoprecipitation (RIP). The 493–644 nt region of miR503HG and the RRM region of hnRNPA2B1 were deemed responsible for this interaction. These regions were discovered through a series of deletion experiments that removed either highly structured portions of the lncRNA or known functional domains of the protein. Such deletion mutants have been very useful for the discovery of RNA binding domains in proteins since it seems that, especially in the case of lncRNA, sequence tends to not be critical for specific binding, but structure (19). Patient overall survival was shown to be better with low presence of the hnRNPA2B1 protein indicating an inverse relationship between miR503HG lncRNA expression and hnRNPA2B1 protein presence and potentially suppressive role for miR503HG against hnRNPA2B1. When assessing 22 HCC tissues, this relationship was confirmed. Wang et al. hypothesized that miR503HG is causing the destabilization of the hnRNPA2B1 protein and to test this hypothesis, performed an experiment overexpressing miR503HG and using a proteasome inhibitor, MG132, to prevent any potential degradation of hnRNPA2B1 by miR503HG through proteasomes. It was revealed that in the presence of MG132, hnRNPA2B1 protein was present at wild-type levels even though miR503HG was present in high levels. Overexpression of miR503HG also led to an increase in the ubiquitination of hnRNPA2B1 providing further evidence that miR503HG was downregulating hnRNPA2B1. These experiments demonstrated that miR503HG and hnRNPA2B1 form a complex, and through this complex miR503HG is able to trigger the ubiquitination of hnRNPA2B1 and destabilize the protein for downregulation.
Finally, Wang et al. chose to go one-step further and deciphered through which signaling pathway lncRNA miR503HG and hnRNPA2B1 were involved in suppressing HCC metastasis. Taking advantage of signaling pathway reporter vectors, it was discovered that when hnRNPA2B1 is knocked-down, the NF-κB pathway was the most significantly downregulated pathway. After thoroughly investigating an exhaustive set of combinations of knockdown and overexpression of miR503HG and hnRNPA2B1, it was established that miR503HG was mediating HCC cell migration through hnRNPA2B1 and the NF-κB pathway. These sets of experiments went so far as to uncover the precise mRNA that hnRNPA2B1 were acting upon (p52 and p65) and downstream genes (c-myc, EZH2, cox2, and VCAM1) that are affected when miR503HG is present. These genes are all key factors in the NF-κB pathway leading to metastasis. The mRNA of the NF-κB subunits, p52 and p65, and the downstream genes are downregulated in the presence of miR503HG. It was reasonably speculated that formation of the hnRNPA2B1-miR503HG complex prevents the attachment of hnRNPA2B1 to the mRNA of p52 and p65 causing the mRNA destabilization and inhibition of the NF-κB pathway (Figure 1). Regulation of mRNA through lncRNA-protein complexes have been proposed previously (18). In a study by Lan et al., it was demonstrated that the lncRNA lnc-HC forms a complex with hnRNPA2B1 to bind and downregulate the mRNA of Cyp7a1 and Abca1. This posttranscriptional regulation aids in the negative regulation of cholesterol metabolism in hepatocytes. This is a slightly different mechanism from the model proposed by Wang et al.; however, it is another example of lncRNA-mediated regulation of hnRNPA2B1. It would be also informative to see if the mRNA of p52 and p65 precipitate with the hnRNPA2B1 protein in a RIP or RNA pull-down assay.
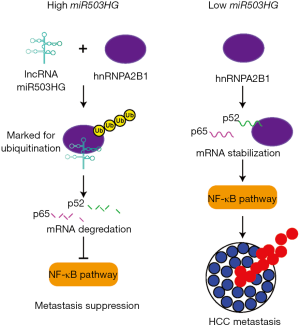
As a whole, Wang et al. were able to demonstrate that miR503HG suppresses metastasis in HCC through binding to the heterogeneous nuclear ribonuclear protein hnRNPA2B1. This study provides a step-step experimental procedure to follow when studying lncRNA in cancer. Wang et al. conducted a comprehensive investigation starting from discovery of lncRNA dysregulation all the way to working out its mechanism of influence in HCC progression. This information can now be fully taken advantage of in the clinic to study how miR503HG can be exploited in the treatment of HCC. Another recent study by Huang et al. in large-cell lymphoma also investigated miR503HG lncRNA and revealed its involvement in the mechanism of proliferation in this cancer (20). This study provides even further support of the important role miR503HG plays in cancer progression not only in HCC, but in other cancers as well. The lncRNA miR503HG potentially could be involved in the metastasis of other cancers through the same mechanism presented by Wang et al. and should be investigated. It would also be prudent to study whether miR503HG can predict the effectiveness of certain treatment modalities in patients. Since Wang et al. already demonstrated its usefulness as a biomarker to predict probability of recurrence after surgery and overall survival (15). Therefore, there is a good chance that it may be a helpful predictor for other treatments such as sorafenib, lenvatinib, and regorafenib (12,14).
This comprehensive study by Wang et al. is a strong case for the involvement of miR503HG in HCC metastasis and for its use as a biomarker for overall survival and recurrence. It has also provided a platform for how lncRNA function can be investigated. Further studies of miR503HG and other lncRNA mechanisms in cancer can help to provide a better picture of cancer genetics. With a better understanding of cancer genetics, treatments that can take advantage of lncRNA mechanisms can be developed.
Acknowledgments
Funding: The authors would like to thank the GAANN Fellowship (P200A120207) from the US Department of Education (to H.M.K.).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Rongrong Gao (Department of Cardiology, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.08.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. [Crossref] [PubMed]
- Perry RB, Ulitsky I. The functions of long noncoding RNAs in development and stem cells. Development 2016;143:3882-94. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300-7. [Crossref] [PubMed]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012;9:703-19. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Perez DS, Hoage TR, Pritchett JR, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet 2008;17:642-55. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Bertuccio P, Turati F, Carioli G, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol 2017;67:302-9. [Crossref] [PubMed]
- Aldrighetti L, Pulitanò C, Catena M, et al. Liver resection with portal vein thrombectomy for hepatocellular carcinoma with vascular invasion. Ann Surg Oncol 2009;16:1254. [Crossref] [PubMed]
- Balogh J, Victor D 3rd, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma 2016;3:41-53. [Crossref] [PubMed]
- Zheng C, Liu X, Chen L, et al. lncRNAs as prognostic molecular biomarkers in hepatocellular carcinoma: a systematic review and meta-analysis. Oncotarget 2017;8:59638-47. [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Wang H, Liang L, Dong Q, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics 2018;8:2814-29. [Crossref] [PubMed]
- Xiao Y, Tian Q, He J, et al. MiR-503 inhibits hepatocellular carcinoma cell growth via inhibition of insulin-like growth factor 1 receptor. Onco Targets Ther 2016;9:3535-44. [PubMed]
- Hasegawa Y, Brockdorff N, Kawano S, et al. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 2010;19:469-76. [Crossref] [PubMed]
- Lan X, Yan J, Ren J, et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology 2016;64:58-72. [Crossref] [PubMed]
- Saldaña-Meyer R, González-Buendía E, Guerrero G, et al. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev 2014;28:723-34. [Crossref] [PubMed]
- Huang PS, Chung IH, Lin YH, et al. The Long Non-Coding RNA MIR503HG Enhances Proliferation of Human ALK-Negative Anaplastic Large-Cell Lymphoma. Int J Mol Sci 2018;19. [PubMed]
Cite this article as: Karner HM, Sun S. LncRNA miR503HG is a new player in hepatocellular carcinoma metastasis. Non-coding RNA Investig 2018;2:51.


