
MicroRNA regulation of CD8+ T cell responses
Introduction
The vertebrate immune system is a finely tuned and remarkably flexible instrument of host defense. A combination of innate pathogen associated molecular pattern recognition and the astonishing diversity and specificity of adaptive immunoreceptors provides protection from omnipresent and ever-changing pathological insults. From viruses to bacteria to eukaryotes to cancer, the diversity and abundance of potential threats that we are constantly encountering is a testament to our immune system’s protective capacity.
CD8+ T cells are adaptive immune cells with an exceptional ability to specifically recognize and kill cells presenting foreign antigens (1). Each CD8+ T cell expresses just one of over a thousand trillion possible versions of the T cell receptor (TCR) with a unique specificity for an antigen that consists of a short peptide presented on class I MHC molecules by potential target cells (2). Upon encountering their cognate antigen, naïve CD8+ T cells become activated, undergo several rounds of cell division thereby generating clones of cells with the same receptor specificity, and differentiate to adopt a diverse multitude of specialized behaviors depending on the context of their activation (1). The factors that govern these cell fates include the abundance of peptide-MHC antigen, TCR affinity for the antigen, the presence or absence of co-stimulatory signals, the local cytokine milieu, and cell-intrinsic factors such as transcription factors and epigenetic regulators of gene expression.
Among the cell-intrinsic factors that regulate T cell differentiation are microRNAs (miRNAs). miRNAs are short (21–24 nucleotide) noncoding RNAs that post-transcriptionally regulate target genes through interaction with their corresponding mRNAs (3). Functional expression of miRNAs is a complex process regulated by machinery that are regulated themselves, to an extent, by miRNAs (4). miRNAs are transcribed (mainly by RNA polymerase II) as long primary transcripts (pri-miRNAs) containing a hairpin structure that contains the mature miRNA sequence. These hairpins are cropped out of pri-miRNAs by the Microprocessor complex, consisting of the RNA binding protein DGCR8 and the RNase III family endoribonuclease Drosha, liberating precursor miRNAs (pre-miRNAs). Pre-miRNAs are exported from the nucleus via exportin 5 where they become accessible to a second RNase III, Dicer, which removes the pre-miRNAs hairpin loop, generating a short RNA duplex. One strand of this duplex becomes a mature miRNA upon its loading into an Argonaute (AGO) protein, forming a miRNA-induced silencing complex (miRISC) (3,4). The miRNA provides specificity to the miRISC, guiding AGO-mediated repression of target mRNAs via Watson-Crick base pairing between the miRNA “seed” sequence and “seed-match” sites found mainly within 3'UTRs. In this manner, miRNAs facilitate translational repression as well as deadenylation and degradation of their target mRNAs.
Adding to the complexity of miRNA regulation, the expression of miRNAs, their target genes, and the machinery required for miRNA function varies between cell types and differentiation states. Therefore, the role of a particular miRNA in one cell type can be dramatically different than its role in another. Depending on the expression level of a particular miRNA and a target RNA, the miRNA may exert little to no effect on protein abundance, tune protein abundance to appropriate levels, or even reduce protein abundance beyond a threshold necessary for effective function in the cell (5). These properties endow miRNAs with the ability to confer robustness to biological processes (6), and to buffer noisy and stochastic transcription that can be leaky and occur in bursts (7-9). Mathematical modeling supported by single-cell reporter assays confirmed a broad role for miRNAs in reducing noise in weakly expressed genes, while surprisingly increasing noise in highly expressed genes (10).
Like transcription factors, miRNAs mediate their biological functions through the regulation of networks of target genes. The magnitude of direct miRNA repression of an individual target mRNA and its corresponding protein is modest, almost never exceeding a two-fold effect. Nevertheless, individual miRNA:target interactions are biologically relevant, as suggested by the evolutionary conservation of many miRNA binding sequences, and supported by naturally occurring variants in miRNA binding sites that alter physiology or confer risk for pathology (11-13). In a few cases, the in vivo requirement for an individual miRNA:target interaction has been interrogated experimentally by mutating a binding sequence by gene targeting in mice (14-17). Each of these mutations was sufficient to produce a profound phenotype, yet none of them could account for the full effect of removing the miRNA entirely. Indeed, all of them involve mutating a single binding site for the same miRNA (miR-155) in different target genes.
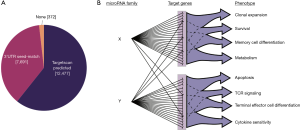
Exemplifying the broad nature of miRNA targeting, algorithms that take into account the conservation of miRNA seed-matches within 3'UTRs predict that over 60 percent of all RefSeq annotated genes can be regulated by one or more miRNAs (11). Biochemical methods that detect miRISC occupancy on mRNAs, such as AGO2 high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (AGO2 HITS-CLIP or AHC) (18) and differential AHC, in which AHC data from cells sufficient or deficient for a particular miRNA are compared (19), generally support these predictions, but indicate that they likely underestimate the extent of miRNA:target interactions. Including all mRNAs with 3'UTR seed-matches, regardless of conservation, expands the list of predicted miRNA targets to include over 98 percent of all RefSeq annotated genes (Figure 1A). Prediction based on sequence alone is not sufficient evidence that targeting occurs. However, it can guide biochemical analyses of miRNA binding to transcripts, as well as expression analyses that measure the context-specific functional effects of miRNAs. Comparative measurement of nascent and mature mRNA levels and quantitative proteomics showed that only 99 targets were stabilized by miR-144/451 deficiency in erythroblasts (20). AHC supported these findings by demonstrating that for most transcripts with miR-144/451-dependent AGO2 binding, there was no appreciable stabilization in miR-144/451 deficient erythroblasts. Further work will be necessary to determine the generalizability of these findings among other miRNAs in different contexts. Importantly, a single RNA can be targeted by multiple miRNAs, and each miRNA influences the expression of tens to hundreds of direct target RNAs (Figure 1B). These targets often include sets of genes that function in common biological pathways, providing opportunities for miRNAs to produce additive phenotypic effects, and to control multiple potential limiting factors in noisy or dynamic gene expression programs.
miRNAs are required for the normal behavior of almost every vertebrate cell type in which their biogenesis or function has been experimentally disabled. In this review, we focus on the regulatory mechanisms that govern CD8+ T cell fate and function within immune responses, emphasizing the roles of miRNAs in controlling or modulating these mechanisms. Additionally, we provide commentary on emergent principles of miRNA regulation.
miRNA regulation of CD8+ T cell function
In response to activation by their cognate antigen, T cells adopt gene expression profiles conducive to rapid proliferation and the deployment of effector functions. While many of these changes occur at the transcriptional level, as much as 50 percent are mediated post-transcriptionally (21). miRNAs also exhibit profound changes in abundance in response to T cell activation owing at least in part to the rapid turnover of miRNA processing machinery and Argonaute proteins (22,23). miR-16, miR-142-3p, miR-150, miR-142-5p, miR-15b, and let-7f are the most abundantly expressed miRNAs in naïve CD8 T cells and all of these miRNAs are down-regulated with in vitro activation (24). Globally, the majority of miRNAs are immediately down-regulated in response to T cell activation (23). However, some, such as miR-155, are transcriptionally upregulated and increase in abundance during T cell activation (25-27).
Essential to an effective cytotoxic T cell response is the proliferation and accumulation of sufficient quantities of antigen-specific cells capable of killing infected cells and cancer. CD8+ T cells lacking miR-155 fail to appropriately expand in response to LCMV infection (28). In fact, in the absence of miR-155, there is a ten-fold reduction in antigen-specific CD8+ T cell accumulation (29) and this appears to be driven by miR-155 effects on both proliferation and survival (30). Members of the miR-17~92 cluster of miRNAs are also up-regulated in response to CD8+ T cell activation in vivo (31,32). Consistent with previous reports describing lymphoproliferative disease resulting from overexpression of the miR-17~92 cluster (33), the proliferative capacity of antigen-specific CD8+ T cells is diminished in the absence of miR-17~92 (32). miR-17~92 has been demonstrated to directly target the tumor suppressor PTEN and the pro-apoptotic protein Bim, providing at least two targets with shared functionality by which miR-17~92 may act. miR-15/16 has been shown to directly target a network of cell cycle and survival associated genes including Ccne1 and Bcl2 (34-36). Consistent with these observations, deletion of miR-15/16 results in increased proliferative capacity and survival among antigen-specific CD8+ T cells (36,37).
Highlighting the importance of miRNAs in restraining CD8+ T cell effector function, Dicer-deficient CD8+ T cells exhibit increased production of perforin, granzyme B, and interferon-gamma (IFN-γ) (38). T cell migration out of central lymphoid organs is a critical component to effector responses and is mediated by surface expression of S1P1 (39), which is inhibited by CD69 (40). Dicer-deficient CD8+ T cells are defective in their ability to migrate out of central lymphoid organs and fail to accumulate in response to infection, likely due to retained surface CD69 expression (41). Several miRNAs that participate in this restraint are downregulated upon T cell activation, linking activating signals to diminishment of a miRNA barrier to T cell differentiation and acquisition of effector functions. Naïve T cells strongly express several members of the let-7 family of miRNAs. Inhibiting let-7 production by overexpression of the regulatory RNA binding protein LIN-28 leads to increased baseline CD8+ T cell proliferation (42). Conversely, let-7 overexpression inhibited antigen-specific T cell clonal expansion and effector function. Retroviral over-expression of pri-miRNAs revealed that miR-139-3p can lead to downregulation of perforin and the transcription factor EOMES, while miR-150 indirectly regulated expression of the high affinity interleukin (IL)-2 receptor, CD25 (38). miR-29 is also down-regulated in response to T cell activation, and is capable of regulating the expression of IFN-γ both directly (43) and indirectly by targeting the transcription factors, T-BET and EOMES (9).
On the other hand, miR-155 deficiency in CD8+ T cells results in reduced cytotoxicity (28) and effector cytokine production (29). In addition to its role in these effector functions, miR-155 enhances the responsiveness of CD8+ T cells to the homeostatic cytokines, IL-7 and IL-15 (44) as well as IL-2 (30). Socs1 knockdown rescues the decreased IL-2 responsiveness of miR-155 deficient T cells. Additionally, Socs1 knockdown in CD8+ T cells phenocopies the tumor-protective effect of miR-155 over-expression. However, while miR-155 plays a critical role in the responsiveness of CD8+ T cells in both acute and chronic infection models, target site mutation indicated that Socs1 repression by miR-155 is only sufficient to mediate a measurable effect in chronic models (17). In addition to Socs1, miR-155 represses Ship1 and Ptpn2, two other negative regulators of AKT and STAT5 signaling (44), among many other direct target mRNAs (19). Overexpression of miR-155 leads to a remarkably improved anti-tumor response by tumor-specific CD8+ T cells (30) suggesting that this approach may be beneficial in adoptive T cell therapies for cancer. The carefully dissected biology of miR-155 in T cell responses provides an edifying example of how miRNAs mediate pleiotropic effects through regulation of a network of target genes.
miRNA regulation of CD8+ T cell memory
In response to infection or cancer, antigen-specific CD8+ T cells may take on the properties of terminally differentiated effector (TE) cells armed with the ability to find and kill infected or abnormal cells, or those of memory precursor (MP) cells capable of persisting long after antigen clearance to provide protection from future encounters. In mice, TE and MP cells can be distinguished early in the immune response based on their surface expression of killer cell lectin like receptor G1 (KLRG1) (45) or the IL-7 receptor alpha chain, CD127 (46), respectively.
A single naïve CD8+ T cell has the potential to give rise to both TE and MP cells (47-49). Asymmetric cell division occurs during clonal selection of antigen-specific T cells, providing one plausible mechanism by which a single antigen-specific T cell could give rise to daughters with differing predetermined lineage fates (50). Noisy expression of lineage-determining factors may also promote stochasticity in these lineage decisions. Responding T cells exhibit a great deal of heterogeneity with respect to proliferative capacity, cytokine production, and the expression of KLRG1 and CD127, and these properties can be intrinsically biased by the TCR even as they are regulated by external cues (51).
No single master transcription factor regulates the differentiation of TE and MP cells, but many contributing factors have been identified. Positive regulators of MP cell differentiation include EOMES (52), TCF1 (53), ID3 (54,55), BCL-6 (56), STAT3 (56), FOXO1 (57,58), BACH2 (59), and MYB (60). Those found to bias cells towards TE fate include T-BET (45,61), BLIMP-1 (62,63), ID2 (50), STAT4 (64), and ZEB2 (65,66). Given that most of these lineage-biasing transcription factors only differ by approximately two-fold in their expression across TE and MP populations, it is likely that the fate of an activated CD8+ T cell is driven by the integration of their effects on downstream target gene networks (50).
Among memory CD8+ T cells, there is a great deal of further heterogeneity. T central memory (Tcm) cells, marked by high expression of CD127, CD62L, CD27, CXCR3, and CCR7, are more proliferative in response to antigen re-challenge. In addition, they tend to exhibit polyfunctionality with respect to effector cytokine secretion, producing IL-2, IFN-γ, and TNF. Conversely, they tend to express lower levels of the cytotoxin granzyme B. Due to their expression of the homing molecules CD62L, CXCR3, and CCR7, Tcm cells circulate through secondary lymphoid organs (SLOs), enhancing their probability of encountering antigen presenting cells (APCs) displaying their cognate antigen. CD27 is a member of the tumor necrosis factor receptor superfamily that acts as a co-stimulatory molecule on Tcm cells, but is absent from TE populations (67). Tcm cells provide superior anti-tumor immunity (68,69) and protection against re-infection with virus (70,71). T effector memory (Tem) populations express high levels of CD127, but little or no CD62L, CXCR3, CD27, and CCR7. Instead, they often express chemotactic and adhesion molecules that allow entry into peripheral, non-lymphoid tissues while excluding them from SLOs. Functionally, Tem cells have enhanced killing capacity due to high expression of the inflammatory cytokines IFN-γ and TNF, as well as the cytotoxic molecules, perforin and granzyme B.
Defining the roles of miRNAs in TE and MP regulation is an active area of research. Several individual miRNAs, or families of miRNAs with identical seed sequences, are known to promote TE or MP differentiation. For example, we recently found that the abundant miR-15/16 family restricts the accumulation of MP cells (36). T cells lacking miR-15/16 exhibit early and sustained increases in MP cell production during the course of viral infection. In wildtype T cells, miR-15/16 bind and repress the expression of hundreds of target genes. Among these targets are a sizable network of memory cell associated genes, including Il7r, which encodes CD127. CD127 is required for the long-term survival of memory cells (72), and IL-7 availability can limit memory cell formation (73). Thus, miR-15/16 may restrict MP differentiation, proliferation and survival in part by tuning their expression of CD127. However, even transgenic over-expression of CD127 is insufficient to enforce memory cell differentiation (72), consistent with the expectation that others among the large number of potentially relevant miR-15/16 targets contribute to this phenotype (36). Of note, transfection of CD8+ T cells with miR-15b mimics led to reduced apoptosis in response to stimulation with anti-CD3 in vitro and was attributed to the down-regulation of the programmed cell death-mediator, DEDD (74). These seemingly conflicting findings among in vivo and in vitro systems highlight the context-dependent nature of miRNA biology in T cells.
At the lower limits of gene expression, the fine tuning function of miRNAs can effectively enforce the silencing of genes with very low or leaky transcription (75). For example, miR-29 regulates IFN-γ production by Dicer-deficient CD4 T cells in part by silencing Eomes, a gene that can co-opt a transcriptional program usually enacted by another miR-29 target, T-BET (9). miR-29 overexpression in CD8 T cells reduces KLRG1+ TE cell production while boosting the frequency of MP cells (9). miR-15/16 may help to enforce silencing of CD127 expression in TE cells, providing robustness to the restriction of IL-7 responsiveness to MP cells. CD127 is maintained at low levels in TE cells via GFI-1 mediated transcriptional silencing (76).
miR-155 increases CD8+ T cell sensitivity to the common gamma chain receptor signaling cytokines, IL-7, IL-15, and IL-2 (30,44). As such, one might predict miR-155 to be a positive regulator of MP cell accumulation. Yet miR-155 is down-regulated in response to in vitro culture of CD8+ T cells with IL-15, and miR-155 deficiency boosts the frequency of CD127+ CD62L+ IL-2 producing MP cells in response to infection with murid herpesvirus (77). In LCMV infection, miR-155 deficiency disrupts the formation of both MP and TE populations, while miR-155 over-expression enhances accumulation of Tem cells (78). Similar observations were reported by several groups, though their attribution of these phenotypes to direct miR-155 target genes varied. SHIP-1 was particularly implicated for its role in the negative regulation of AKT phosphorylation and T-BET expression (28). However, once again it appears likely that miR-155 acts through a network of targets with additive or synergistic cell intrinsic effects on antiviral CD8+ T cell responses (29).
The transcription factors ZEB1 and ZEB2 play reciprocal roles in the promotion of MP and TE cell differentiation respectively (79). ZEB2 is expressed among terminally differentiated cytotoxic T cells in a T-bet-dependent manner and plays a critical role in the suppression of MP associated genes while positively regulating TE-associated genes (65,66). Although the miR-200 family of miRNAs have been shown to negatively regulate both Zeb1 and Zeb2 in the context of epithelial differentiation (80), only Zeb2 mRNA appears to interact with, and be efficiently targeted by, miR-200 family miRNAs in T cells. Consistent with these findings, miR-200 was essential for normal memory CD8+ T cell differentiation (79). This study documents the importance of cellular context to miRNA-target interactions, further highlighting the utility of empirical target identification by co-immunoprecipitation with AGO proteins.
Metabolism plays a critical role in the proliferation and differentiation of CD8+ T cells. Inhibition of mTOR, for example, enhances memory cell persistence through the switch from glycolysis to fatty acid metabolism (81,82) and through the regulation of T-BET and EOMES (83). Over-expression of the miR-17~92 cluster of miRNAs enhances TE differentiation, while miR-17~92 deficient CD8+ T cells are biased towards a MP phenotype (32). miR-17~92 regulates PI3K-AKT-mTOR signaling in T cells, and many other cell types, by targeting PTEN, a tightly tuned lipid and protein phosphatase that counteracts this signaling axis. These results do not rule out other targets as players. In fact, miR-17~92 also downregulates additional negative regulators of mTOR (e.g., Pdcd1, Btla, and Fcgr2b), though direct targeting and functional relevance of these putative targets has not been confirmed (31). The importance of functional testing of targets and consideration of the full suite of targets of a miRNA is highlighted by the case of mir-15/16, which regulates both mTOR and Rictor (37,84), but nevertheless restricts, rather than enhances, memory cell differentiation.
In human CD8+ T cells, miR-143 overexpression enhanced the production of cells expressing the memory markers CD127, CD27 and CD28, whereas miR-143 inhibition reduced them (85). These effects were attributed to the miR-143 target Glut1, whose knockdown produced a phenotype consistent with that produced by miR-143 overexpression. The rate-limiting glycolysis enzyme HK2 was also down-regulated upon miR-143 overexpression (85), consistent with previous reports of direct targeting by miR-143 (86). Thus miR-143 coordinates the expression of at least two key targets in a pathway (glycolytic metabolism) critical to the regulation of CD8+ T cell responses.
The roles of miRNAs in the transitions between memory cell states remains poorly understood. Transfer of CD62Lhi and CD62Llo antigen-specific CD8+ T cells supports a model of linear transition from naïve to Tem to Tcm cell identities (70). And KLRG1+ CD127+ Tem cells can lose KLRG1 expression and seed Tcm cell compartments in a Bach2-dependent manner (87). While over-expression of miR-150 decreases CD127+ CXCR3+ Tcm populations, subsequent reduced expression at later timepoints leads to an increase in Tcm cells (88). In some systems, miR-150 deficient memory T cells remained defective in recall responses and cytolytic function (89). Overexpression of the miR-150 target MYB was sufficient to partially rescue some of the memory cell defects associated with miR-150 over-expression (88). This may be at least in part attributable to the indispensable role of MYB in regulating CD8+ T cell stemness, memory, and polyfunctionality (60). Another study confirmed that miR-150 restrains memory CD8+ T cell differentiation but attributed these effects to another direct target, Foxo1, a transcription factor that drives memory cell differentiation through induction of TCF1 (90). The established miR-150 target network contains several other functionally relevant target genes, including Trp53 (91), Slc2a1 (92), Mtor (93), Ptrx7 (94), Egr2 (95), in addition to indirect effects on CD25 expression (38).
miRNA regulation of CD8+ T cell exhaustion
Effective memory generation requires the clearance of the pathogen or tumor. Persistent antigen exposure induces CD8+ T cell “exhaustion”, characterized by upregulation of inhibitory receptors including PD1, LAG3, and CTLA4, concomitant with reduced proliferation capacity, effector function and cell survival (96). Understanding the drivers and maintainers of T cell exhaustion is especially pressing in the context of tumor immunology. Over the two past decades, it has become evident that the reversal of T cell exhaustion can unleash existing tumor-specific cytotoxic T cells to attack and kill cancerous cells. Blocking inhibitory receptors can reverse exhaustion and induce productive antiviral and antitumor immunity (97-99). PD-1 blockade only temporarily reinvigorates exhausted CD8+ T cells if the causative antigen is not cleared, indicating that targeting these surface receptors alone may be insufficient for many immunotherapies (100). Effective strategies for durably reprogramming exhausted T cells may improve existing and developing immunotherapies for cancer.
A growing number of transcription factors have been implicated in T cell exhaustion including T-BET, EOMES, Spry2, BLIMP-1, VHL, FOXO1, IRF, BATF, and NFATC1 (96,101) and more recently, the NR4A family members, NR4A1, NR4A2, and NR4A3 (102). Interestingly, many of these transcription factors are also critical to functional effector and memory CD8+ T cells, suggesting a complexity to the drivers of exhaustion that remains to be fully understood. For example, IRF4 and BATF are essential for CD8+ T cell effector function during infection with LCMV (103), but they can also elevate PD-1 expression and repress TCF1, thereby promoting exhaustion while inhibiting memory formation (101).
miRNAs also play a role in CD8+ T cell exhaustion. In the context of cancer, tumor-derived TGF-β can lead to elevation of miR-23a expression, thereby downregulating BLIMP-1 and a loss of effector function (104). Functional inhibition of miR-23a led to improved effector function and a more durable response to established tumors. In response to chronic infection with LCMV clone 13, miR-31 deficient CD8+ T cells express reduced levels of exhaustion markers and retain characteristics of effector cells, including production of cytotoxins and cytokines (105). Mice lacking miR-31 expression only in T cells were protected from the wasting associated with chronic infection and harbored lower viral titers. miR-155 overexpression enhances the persistence of exhausted CD8+ T cells during chronic infection (106). Uncoupling persistence from effector function, miR-155 overexpression fails to restore cytokine production and cytotoxic potential, and actually increases inhibitory receptor expression in these cells. Unlike memory cells, exhausted CD8+ T cells do not require the homeostatic cytokines IL-7 and IL-15 to persist and instead rely on constant exposure to their cognate antigen (107). Thus, miR-155’s ability to alter CD8+ T cell sensitivity to common gamma chain cytokine receptor signals is unlikely to be responsible for increased persistence of miR-155-overexpressing exhausted T cells. Conversely, in the chronic setting of cancer, miR-155 overexpression delays CD8+ T cell contraction, prolongs cytokine production, and increases their sensitivity to common gamma chain cytokines (44). miR-31 may reduce exhaustion in part by increasing CD8+ T cell sensitivity to type I IFNs (105). However, miR-155 decreases IFN sensitivity (29), so this is also unlikely to play a role in the persistence of miR-155 overexpressing exhausted T cells. The varied effects of miR-155 on T cells responses to acute and chronic infection as well as cancer highlight the context specific nature of miRNAs regulation of immunity.
Perspective
miRNAs are important regulators of CD8+ T cell function in host defense, infection, and cancer immunosurveillance. They regulate almost every aspect of CD8+ T cell behavior from survival and proliferation, to the acquisition and deployment of effector functions, to fate decisions that dictate the formation of immunological memory and tolerance. Individual miRNA-target interactions can have profound impacts on cell behavior, but mounting evidence indicates that miRNAs mediate context-specific biological effects by binding and tuning the expression of large networks of target genes. There remains much to learn about miRNA regulation in CD8+ T cells, and perhaps even more to learn about CD8+ T cell programming from the study of miRNAs. While somewhat useful in the validation of target sites, assays that remove the 3'UTRs of putative target genes from their endogenous context (e.g., luciferase assays) suffer from a potential for both false-positive and false-negative findings. Furthermore, the singling out of individual targets and the highlighting of the capacity for their 3'UTRs to be regulated by a particular miRNA draws attention away from the underlying nature of miRNAs as network regulators of gene circuits. The field of miRNA biology will benefit greatly from a more systematic identification of miRNA targets within each cellular context and formulating a catalog of the pathways and gene sets involved. To this end, future studies should expand on efforts to map the full target repertoire of functionally relevant miRNAs using a combination of bioinformatics, biochemistry (AHC), and analysis of endogenous miRNA effects on gene expression in biologically relevant contexts. These target-agnostic approaches invite the discovery of novel targets and will lead to better assessment of how their associated pathways integrate with phenotype.
Acknowledgments
Funding: This work was supported by the National Institute of Health (HL109102, HL107202), National Science Foundation Graduate Research Fellowship Program (2015202541 to JD Gagnon) and the Sandler Asthma Basic Research Center.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2019.07.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity 2011;35:161-8. [Crossref] [PubMed]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature 1988;334:395-402. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. [Crossref] [PubMed]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5:396-400. [Crossref] [PubMed]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell 2012;149:515-24. [Crossref] [PubMed]
- Raj A, Peskin CS, Tranchina D, et al. Stochastic mRNA synthesis in mammalian cells. PLoS Biol 2006;4:e309 [Crossref] [PubMed]
- Blake WJ, Balázsi G, Kohanski MA, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 2006;24:853-65. [Crossref] [PubMed]
- Steiner DF, Thomas MF, Hu JK, et al. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 2011;35:169-81. [Crossref] [PubMed]
- Schmiedel JM, Klemm SL, Zheng Y, et al. Gene expression. MicroRNA control of protein expression noise. Science 2015;348:128-32. [Crossref] [PubMed]
- Friedman RC, Farh KKH, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [Crossref] [PubMed]
- Clop A, Marcq F, Takeda H, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 2006;38:813-8. [Crossref] [PubMed]
- Steri M, Orrù V, Idda ML, et al. Overexpression of the Cytokine BAFF and Autoimmunity Risk. N Engl J Med 2017;376:1615-26. [Crossref] [PubMed]
- Teng G, Hakimpour P, Landgraf P, et al. MicroRNA-155 Is a Negative Regulator of Activation-Induced Cytidine Deaminase. Immunity 2008;28:621-9. [Crossref] [PubMed]
- Dorsett Y, McBride KM, Jankovic M, et al. MicroRNA-155 Suppresses Activation-Induced Cytidine Deaminase-Mediated Myc-Igh Translocation. Immunity 2008;28:630-8. [Crossref] [PubMed]
- Lu D, Nakagawa R, Lazzaro S, et al. The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J Exp Med 2014;211:2183-98. [Crossref] [PubMed]
- Lu LF, Gasteiger G, Yu IS, et al. A Single miRNA-mRNA Interaction Affects the Immune Response in a Context- and Cell-Type-Specific Manner. Immunity 2015;43:52-64. [Crossref] [PubMed]
- Chi SW, Zang JB, Mele A, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009;460:479-86. [Crossref] [PubMed]
- Loeb GB, Khan AA, Canner D, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell 2012;48:760-70. [Crossref] [PubMed]
- Xu P, Palmer LE, Lechauve C, et al. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood 2019;133:2518-28. [Crossref] [PubMed]
- Cheadle C, Fan J, Cho-Chung YS, et al. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics 2005;6:75. [Crossref] [PubMed]
- Bronevetsky Y, Ansel KM. Regulation of miRNA biogenesis and turnover in the immune system. Immunol Rev 2013;253:304-16. [Crossref] [PubMed]
- Bronevetsky Y, Villarino AV, Eisley CJ, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med 2013;210:417-32. [Crossref] [PubMed]
- Wu H, Neilson JR, Kumar P, et al. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One 2007;2:e1020 [Crossref] [PubMed]
- Haasch D, Chen YW, Reilly RM, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol 2002;217:78-86. [Crossref] [PubMed]
- Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007;316:608-11. [Crossref] [PubMed]
- Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science 2007;316:604-8. [Crossref] [PubMed]
- Lind EF, Elford AR, Ohashi PS. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol 2013;190:1210-6. [Crossref] [PubMed]
- Gracias DT, Stelekati E, Hope JL, et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol 2013;14:593-602. [Crossref] [PubMed]
- Dudda JC, Salaun B, Ji Y, et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity 2013;38:742-53. [Crossref] [PubMed]
- Wu T, Wieland A, Araki K, et al. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proc Natl Acad Sci USA 2012;109:9965-70. [Crossref] [PubMed]
- Khan AA, Penny LA, Yuzefpolskiy Y, et al. MicroRNA-17~92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood 2013;121:4473-83. [Crossref] [PubMed]
- Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 2008;9:405-14. [Crossref] [PubMed]
- Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 2005;102:13944-9. [Crossref] [PubMed]
- Ofir M, Hacohen D, Ginsberg D. miR-15 and miR-16 Are Direct Transcriptional Targets of E2F1 that Limit E2F-Induced Proliferation by Targeting Cyclin E. Mol Cancer Res 2011;9:440-7. [Crossref] [PubMed]
- Gagnon JD, Kageyama R, Shehata HM, et al. miR-15/16 Restrain Memory T Cell Differentiation, Cell Cycle, and Survival. Cell Rep 2019; [Crossref] [PubMed]
- Yang J, Liu R, Deng Y, et al. MiR-15a/16 deficiency enhances anti-tumor immunity of glioma-infiltrating CD8+ T cells through targeting mTOR. Int J Cancer 2017;141:2082-92. [Crossref] [PubMed]
- Trifari S, Pipkin ME, Bandukwala HS, et al. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci USA 2013;110:18608-13. [Crossref] [PubMed]
- Matloubian M, Lo CG, Cinamon G, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004;427:355-60. [Crossref] [PubMed]
- Shiow LR, Rosen DB, Brdicková N, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 2006;440:540-4. [Crossref] [PubMed]
- Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA 2010;107:21629-34. [Crossref] [PubMed]
- Wells AC, Daniels KA, Angelou CC, et al. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. Elife 2017;6:403. [Crossref] [PubMed]
- Ma F, Xu S, Liu X, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol 2011;12:861-9. [Crossref] [PubMed]
- Ji Y, Wrzesinski C, Yu Z, et al. miR-155 augments CD8+ T-cell antitumor activity in lymphoreplete hosts by enhancing responsiveness to homeostatic γc cytokines. Proc Natl Acad Sci USA 2015;112:476-81. [Crossref] [PubMed]
- Joshi NS, Cui W, Chandele A, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 2007;27:281-95. [Crossref] [PubMed]
- Kaech SM, Tan JT, Wherry EJ, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol 2003;4:1191-8. [Crossref] [PubMed]
- Stemberger C, Huster KM, Koffler M, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity 2007;27:985-97. [Crossref] [PubMed]
- Gerlach C, van Heijst JW, Swart E, et al. One naive T cell, multiple fates in CD8+ T cell differentiation. J Exp Med 2010;207:1235-46. [Crossref] [PubMed]
- Gerlach C, Rohr JC, Perié L, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science 2013;340:635-9. [Crossref] [PubMed]
- Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol 2014;15:1104-15. [Crossref] [PubMed]
- Plumlee CR, Sheridan BS, Cicek BB, et al. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity 2013;39:347-56. [Crossref] [PubMed]
- Banerjee A, Gordon SM, Intlekofer AM, et al. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 2010;185:4988-92. [Crossref] [PubMed]
- Zhou X, Yu S, Zhao DM, et al. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 2010;33:229-40. [Crossref] [PubMed]
- Yang CY, Best JA, Knell J, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol 2011;12:1221-9. [Crossref] [PubMed]
- Ji Y, Pos Z, Rao M, et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol 2011;12:1230-7. [Crossref] [PubMed]
- Cui W, Liu Y, Weinstein JS, et al. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 2011;35:792-805. [Crossref] [PubMed]
- Hess Michelini R, Doedens AL, Goldrath AW, et al. Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med 2013;210:1189-200. [Crossref] [PubMed]
- Kim MV, Ouyang W, Liao W, et al. The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 2013;39:286-97. [Crossref] [PubMed]
- Roychoudhuri R, Clever D, Li P, et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 2016;17:851-60. [Crossref] [PubMed]
- Gautam S, Fioravanti J, Zhu W, et al. The transcription factor c-Myb regulates CD8+ T cell stemness and antitumor immunity. Nat Immunol 2019;20:337-49. [Crossref] [PubMed]
- Intlekofer AM, Takemoto N, Kao C, et al. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med 2007;204:2015-21. [Crossref] [PubMed]
- Kallies A, Xin A, Belz GT, et al. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity 2009;31:283-95. [Crossref] [PubMed]
- Rutishauser RL, Martins GA, Kalachikov S, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 2009;31:296-308. [Crossref] [PubMed]
- Mollo SB, Ingram JT, Kress RL, et al. Virus-specific CD4 and CD8 T cell responses in the absence of Th1-associated transcription factors. J Leukoc Biol 2014;95:705-13. [Crossref] [PubMed]
- Dominguez CX, Amezquita RA, Guan T, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med 2015;212:2041-56. [Crossref] [PubMed]
- Omilusik KD, Best JA, Yu B, et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med 2015;212:2027-39. [Crossref] [PubMed]
- Hintzen RQ, de Jong R, Lens SM, et al. Regulation of CD27 expression on subsets of mature T-lymphocytes. J Immunol 1993;151:2426-35. [PubMed]
- Klebanoff CA, Gattinoni L, Torabi-Parizi P, et al. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc Natl Acad Sci USA 2005;102:9571-6. [Crossref] [PubMed]
- Enamorado M, Iborra S, Priego E, et al. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat Commun 2017;8:16073. [Crossref] [PubMed]
- Wherry EJ, Teichgräber V, Becker TC, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 2003;4:225-34. [Crossref] [PubMed]
- Hikono H, Kohlmeier JE, Takamura S, et al. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med 2007;204:1625-36. [Crossref] [PubMed]
- Hand TW, Morre M, Kaech SM. Expression of IL-7 receptor alpha is necessary but not sufficient for the formation of memory CD8 T cells during viral infection. Proc Natl Acad Sci USA 2007;104:11730-5. [Crossref] [PubMed]
- Nanjappa SG, Walent JH, Morre M, et al. Effects of IL-7 on memory CD8 T cell homeostasis are influenced by the timing of therapy in mice. J Clin Invest 2008;118:1027-39. [PubMed]
- Zhong G, Cheng X, Long H, et al. Dynamically expressed microRNA-15b modulates the activities of CD8+ T lymphocytes in mice with Lewis lung carcinoma. J Transl Med 2013;11:71. [Crossref] [PubMed]
- Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 2009;6:570-5. [Crossref] [PubMed]
- Carrette F, Surh CD. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol 2012;24:209-17. [Crossref] [PubMed]
- Tsai CY, Allie SR, Zhang W, et al. MicroRNA miR-155 affects antiviral effector and effector Memory CD8 T cell differentiation. J Virol 2013;87:2348-51. [Crossref] [PubMed]
- Hope JL, Stairiker CJ, Spantidea PI, et al. The Transcription Factor T-Bet Is Regulated by MicroRNA-155 in Murine Anti-Viral CD8+ T Cells via SHIP-1. Front Immunol 2017;8:1696. [Crossref] [PubMed]
- Guan T, Dominguez CX, Amezquita RA, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8 +T cell fates. J Exp Med 2018;215:1153-68. [Crossref] [PubMed]
- Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep 2010;11:670-7. [Crossref] [PubMed]
- Pearce EL, Walsh MC, Cejas PJ, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 2009;460:103-7. [Crossref] [PubMed]
- Araki K, Turner AP, Shaffer VO, et al. mTOR regulates memory CD8 T-cell differentiation. Nature 2009;460:108-12. [Crossref] [PubMed]
- Rao RR, Li Q, Odunsi K, et al. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity 2010;32:67-78. [Crossref] [PubMed]
- Singh Y, Garden OA, Lang F, et al. MicroRNA-15b/16 Enhances the Induction of Regulatory T Cells by Regulating the Expression of Rictor and mTOR. J Immunol 2015;195:5667-77. [Crossref] [PubMed]
- Zhang T, Zhang Z, Li F, et al. miR-143 Regulates Memory T Cell Differentiation by Reprogramming T Cell Metabolism. J Immunol 2018;201:2165-75. [Crossref] [PubMed]
- Peschiaroli A, Giacobbe A, Formosa A, et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene 2013;32:797-802. [Crossref] [PubMed]
- Herndler-Brandstetter D, Ishigame H, Shinnakasu R, et al. KLRG1+ Effector CD8+ T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity 2018;48:716-8. [Crossref] [PubMed]
- Chen Z, Stelekati E, Kurachi M, et al. miR-150 Regulates Memory CD8 T Cell Differentiation via c-Myb. Cell Rep 2017;20:2584-97. [Crossref] [PubMed]
- Smith NL, Wissink EM, Grimson A, et al. miR-150 Regulates Differentiation and Cytolytic Effector Function in CD8+ T cells. Sci Rep 2015;5:16399. [Crossref] [PubMed]
- Ban YH, Oh SC, Seo SH, et al. miR-150-Mediated Foxo1 Regulation Programs CD8+ T Cell Differentiation. Cell Rep 2017;20:2598-611. [Crossref] [PubMed]
- Zhang N, Wei X, Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett 2013;587:2346-51. [Crossref] [PubMed]
- King BC, Esguerra JLS, Golec E, et al. CD46 Activation Regulates miR-150-Mediated Control of GLUT1 Expression and Cytokine Secretion in Human CD4+ T Cells. J Immunol 2016;196:1636-45. [Crossref] [PubMed]
- Warth SC, Hoefig KP, Hiekel A, et al. Induced miR-99a expression represses Mtor cooperatively with miR-150 to promote regulatory T-cell differentiation. EMBO J 2015;34:1195-213. [Crossref] [PubMed]
- Zhou L, Qi X, Potashkin JA, et al. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3'-untranslated region of the gene that decrease steady-state levels of the transcript. J Biol Chem 2008;283:28274-86. [Crossref] [PubMed]
- Bousquet M, Zhuang G, Meng C, et al. miR-150 blocks MLL-AF9-associated leukemia through oncogene repression. Mol Cancer Res 2013;11:912-22. [Crossref] [PubMed]
- Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 2015;15:486-99. [Crossref] [PubMed]
- Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996;271:1734-6. [Crossref] [PubMed]
- Barber DL, Wherry EJ, Masopust D, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006;439:682-7. [Crossref] [PubMed]
- Blackburn SD, Shin H, Haining WN, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 2009;10:29-37. [Crossref] [PubMed]
- Pauken KE, Sammons MA, Odorizzi PM, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 2016;354:1160-5. [Crossref] [PubMed]
- Man K, Gabriel SS, Liao Y, et al. Transcription Factor IRF4 Promotes CD8+ T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 2017;47:1129-41.e5. [Crossref] [PubMed]
- Chen J, López-Moyado IF, Seo H, et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 2019;567:530-4. [Crossref] [PubMed]
- Grusdat M, McIlwain DR, Xu HC, et al. IRF4 and BATF are critical for CD8+ T-cell function following infection with LCMV. Cell Death Differ 2014;21:1050-60. [Crossref] [PubMed]
- Lin R, Chen L, Chen G, et al. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest 2014;124:5352-67. [Crossref] [PubMed]
- Moffett HF, Cartwright ANR, Kim HJ, et al. The microRNA miR-31 inhibits CD8+ T cell function in chronic viral infection. Nat Immunol 2017;18:791-9. [Crossref] [PubMed]
- Stelekati E, Chen Z, Manne S, et al. Long-Term Persistence of Exhausted CD8 T Cells in Chronic Infection Is Regulated by MicroRNA-155. Cell Rep 2018;23:2142-56. [Crossref] [PubMed]
- Shin H, Blackburn SD, Blattman JN, et al. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med 2007;204:941-9. [Crossref] [PubMed]
Cite this article as: Gagnon JD, Ansel KM. MicroRNA regulation of CD8+ T cell responses. Non-coding RNA Investig 2019;3:24.


