Towards a universal nomenclature standardization for circular RNAs
Circular RNAs (circRNAs) are non-coding RNAs generated by a non-canonical splicing phenomenon, constituting a very dynamic family of molecules with a wide regulatory potential (1). Their ubiquitous presence and functional relevance within cells have been described in the last decade (2), but this subject has recently become a very vibrant and hot scientific topic as stated by the number of peer-reviewed manuscripts published in the last year 2019 (~1,500 papers in 2019 as indexed in PubMed database).
Scientists working on the circRNA field are currently facing the problem of the lack of standardization in the nomenclature of these molecules, which prevents the validation of experimental results and a deeper knowledge about their functional roles. Despite of the efforts of several research groups for the building of central resources to compile circRNA information, the databases are incomplete, not interconnected, constituting isolated islands and all of them have their own nomenclatures to annotate circRNAs. There are databases compiling restricted groups of circRNAs and their functions in the context of functional events or producing species, but only four resources could be considered as general-purpose and resource databases for circRNAs: circBase (3), circBank (4), CIRCpedia (5) and circAtlas (6). circBase was the precursor database and proposed a specific nomenclature for circRNAs which includes the species and a numeric code, whereas circBank and circAtlas use a more friendly annotation considering also the gene symbol of the transcriptional unit responsible for the generation of a circRNA based on the genomic coordinate references from UCSC resources (7). CIRCpedia uses a specific denomination for each circRNA, including the original species and an internal number without the reference to the source gene, as observed in circBase nomenclature (3). Together to these all different nomenclatures, there is an additional layer of complexity motivated by the presence of specialized companies devoted to the commercialization of molecular biology products, that also proposed their own and proprietary annotation for circRNAs, which is only accessible if the interested scientists are customers. Due to this complex scenario, research papers describing screenings or individual characterization of circRNAs are full of different nomenclatures including the already described sources, but also proposing new ones. Under these circumstances, the replication of functional results regarding a specific circRNA or even the simple obtention of its sequence is almost a “mission impossible”.
Due to the scientific background already available, the field of circRNAs is mature enough to be supported on solid foundations, and the existence of a normalized nomenclature in all databases is essential. An excellent illustration about how universal nomenclature can contribute to the scientific progress and data interchange in the field of ncRNAs is the miRbase resource, devoted to the compilation of microRNA (miRNA) data (8). This database has evolved during the last fifteen years adapting itself to the evolution of laboratory techniques used for the characterization of miRNAs and the increasing available data from different species. Current miRbase version is 22.1, and includes information about miRNA sequence, biological origin, relative expression and biological function, based on a standard annotation scheme (8).
Considering the exposed facts, we strongly believe in the need of a standardized annotation for circRNAs, which also might be complemented by methodological rules that should be followed for the scientists within the field. We propose the following recommendations for the annotation and publication of data related with circRNAs:
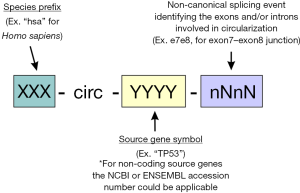
- A possible standardized nomenclature for circRNAs could follow a design which combines the already well established miRbase rules together with the circBank annotation, as illustrated in Figure 1. Following this annotation, circRNA identifications should include the acronym for the producing species, the source transcriptional unit and a representation of the back-splicing event showing the connection between gene structural units (this nomenclature would identify the 5' and 3' donors in back splicing by using a letter, “e” for exon and “i” for intron, followed by the sequential number in the gene structure). The designation of source transcriptional units could be achieved by using the canonical gene symbol for coding genes, and the ENSEMBL or NCBI reference for ncRNA genes (long non-coding RNAs and other non-coding transcriptional units).
- The proposed nomenclature could be easily adopted and implemented by all the already existing resources as circBase and circBank by including an “alias” record.
- Resources devoted to the compilation and storage of circRNA information would require a stronger crosstalk for information exchange, ensuring the proper relationships and equivalences among circRNA entries and annotations. Sequences of circRNAs compiled in databases should be properly indexed including information about the reference genomic assembly, and the gene structural units involved in the non-canonical splicing events.
- The computer software currently used for the detection, quantification and differential expression of circRNAs from high-throughput sequencing data is based on the detection of non-canonical exon-intron junctions, and strongly dependent on the proper indexing of the reference genome. In consequence and for ensuring proper reproducibility and validation of experimental results, scientific publications which describe and analyze the function of circRNAs should also include a precise description of their genomic coordinates, describing the version of the genomic assembly employed for indexing, and the relevant annotation equivalences in all the general-purpose databases. Publishing policies in specialized journals should also consider that non-standard circRNA annotations should be avoided in order to ensure a homogeneous exchange of information among scientists.
- Companies commercializing proprietary technologies for the analysis and screening of circRNAs, should include the normalized circRNA nomenclature for their products and make it available to the general scientific community and not only for their customers. Open information about genomic sequences of circRNAs detected by protected technologies would have no commercial disadvantages for commercial suppliers, a fact that has been already demonstrated in proprietary technologies for the detection of other ncRNAs as miRNAs (9).
Acknowledgments
Funding: This work is supported by COST (European Cooperation in Science and Technology) Action EU-CardioRNA CA17129 and Portuguese Foundation for Science and Technology (FCT) under the framework of the research grant PTDC-MED-GEN-29389-2017.
Footnote
Provenance and Peer Review: This is a free submission. The article did not undergo external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2020.03.01). All authors report grants from Fundação para a Ciência e a Tecnologia (FCT), during the conduct of the study. FJE serves as an unpaid editorial board member of Non-coding RNA Investigation from May 2019 to April 2021.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Santer L, Bär C, Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol Ther 2019;27:1350-63. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666-70. [Crossref] [PubMed]
- Liu M, Wang Q, Shen J, et al. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol 2019;16:899-905. [Crossref] [PubMed]
- Dong R, Ma XK, Li GW, et al. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatics 2018;16:226-33. [Crossref] [PubMed]
- Ji P, Wu W, Chen S, et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep 2019;26:3444-60.e5. [Crossref] [PubMed]
- Haeussler M, Zweig AS, Tyner C, et al. The UCSC genome browser database: 2019 update. Nucleic Acids Res 2019;47:D853-8. [Crossref] [PubMed]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155-62. [Crossref] [PubMed]
- Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 2004;43:13233-41. [Crossref] [PubMed]
Cite this article as: Costa MC, Enguita FJ. Towards a universal nomenclature standardization for circular RNAs. Non-coding RNA Investig 2020;4:2.