
High-precision screen to identify lncRNAs governing specific mitotic stages
The vast majority of human transcripts lack coding potential (1,2). These transcripts are known as noncoding RNAs (ncRNAs) and include many ubiquitous RNAs with housekeeping functions such as ribosomal (r)RNAs and transfer (t)RNAs, and many RNAs with restricted spatiotemporal expression patterns, including long noncoding (lnc)RNAs, micro (mi)RNAs, and piwi-interacting (pi)RNAs, which regulate gene expression programs in a range of cellular processes (3). The study discussed here focuses on lncRNAs, a class of ncRNAs longer than 200 nucleotides that can regulate gene expression on many levels by remodeling chromatin, modulating transcription, affecting posttranslational processes, and altering protein activity (3-5). Through these functions, lncRNAs can affect cell proliferation, differentiation, and apoptosis, thereby contributing to pathologies such as cancer (6,7). Recent reports have highlighted distinct roles of lncRNAs in mitotic cell division (8-11).
Following DNA replication, cells enter a period of division into two daughter cells through mitosis, during which chromosomes condense, orient, and segregate; afterwards, cells undergo cytokinesis, during which the nuclear and cytoplasmic contents divide (12). Chromosome condensation is often tracked by monitoring phosphorylation of histone H3 (PHH3), which marks cells at the G2- to M-phase transition (13). Thus, in a given cell population, the mitotic index (the ratio between the number of mitotic cells and the number of total cells) can be calculated as PHH3-positive cells relative to total cells. Chromosome segregation requires the precise coordination of spindle assembly and polymerization of microtubules (consisting of α- and β-tubulin dimers) that separate sister chromatids toward opposite poles (14). As soon as mitosis ends, a contractile ring cleaves the cytokinetic bridges connecting the two daughter cells to eventually split them. The microtubules that form cytokinetic bridges are often used to detect cytokinetic progression. If these bridges remain attached, daughter cells do not separate and mitosis is halted (15). In light of growing evidence that lncRNAs govern cell division, Stojic and colleagues recently set out to identify lncRNAs affecting cell division using RNA interference (RNAi) and high-content screening (16).
Stojic et al. employed human cervical carcinoma HeLa cells to set up a tour de force screen of small interfering (si)RNAs targeting 2,231 lncRNAs (the human Lincode siRNA library), then assessing mitotic traits with state-of-the-art methods. In the first screen the authors focused on defects of chromosome segregation and cytokinesis by visualizing centrosomes stained for the centromeric protein CEP215 and by visualizing the actin cytoskeleton using phalloidin (17). In the second screen, the authors focused on cells entering the M-phase and the maturation of centrosomes by staining for PHH3 and γ-tubulin, respectively (17). In both screens, Hoechst was used to visualize DNA and α-tubulin was used to assess cytoskeletal structures; in the process, the authors obtained highly informative data and strikingly beautiful images.
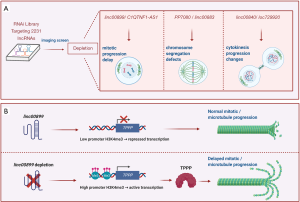
Among the most prominent lncRNAs identified as affecting mitosis, silencing linc00899 and C1QTNF1-AS1 was found to promote mitotic delay, silencing linc00883 and PP7080 increased chromosome segregation errors, silencing linc00840 doubled the cytokinetic cleavage time, and silencing loc729970 shortened cytokinesis (summarized in Figure 1A). The authors further analyzed linc00899 and C1QTNF1-AS1, two spliced and polyadenylated lncRNAs spanning ~1.6 and ~1 kb, respectively. Linc00899 (also known as loc100271722 or ENSG00000231711) lacks a mouse ortholog, but harbors short conserved stretches within exon 1, while C1QTNF1-AS1 (also known as ENSG00000265096) is an antisense RNA transcribed from a locus that encodes the C1Q and TNF-related 1 (C1QTNF1/CTRP1) protein and is conserved between human and mouse.
The authors further confirmed the RNAi-mediated mitotic delay by silencing these lncRNAs using locked-nucleic-acid (LNA) oligomers. Subsequent analysis of differentially expressed genes revealed that depleting linc00899 changed the expression levels of many transcripts, while depleting C1QTNF1-AS1 affected mainly C1QTNF1 mRNA levels. Therefore, the authors focused on linc00899 and specifically on the differentially regulated TPPP/p25 mRNA; the encoded product, TPPP (the ubiquitous tubulin polymerization-promoting protein), blocks mitotic spindle formation by promoting tubulin polymerization (18). Reverse transcription (RT) followed by quantitative (q)PCR analysis and western blot analysis verified a rise in TPPP mRNA and TPPP production when linc00899 was silenced. Double knockdowns of linc00899 and TPPP confirmed that the mitotic delay by linc00899 depletion was mediated through the upregulation of TPPP in HeLa cells. Depletion of linc00899 increased α-tubulin acetylation and long-lived microtubules, mimicking the microtubule-stabilizing function of TPPP (19), and reduced the levels of end-binding 1 (EB1) protein, which associates with the ends of growing microtubules (20). Accordingly, in linc00899-depleted cells, metaphase plates were narrow, with unattached kinetochore pairs predominantly located near the poles. Furthermore, in linc00899-depleted cells, the chromosome bi-orientation in metaphase was altered only modestly, while the reduction in interkinetochore distances (inter-KT) of chromosomes suggested that linc00899 affected the tension to kinetochore pairs in metaphase (21).
To gain insight into how linc00899 might modulate TPPP levels and function, the authors investigated the methylation of the TPPP promoter. Strikingly, silencing linc00899 increased trimethylation of lysine 4 on histone H3 (H3K4me3) at the TPPP promoter (Figure 1B), although there were no sequences complementary to the linc00899 transcript. These findings suggest that an independent ribonucleoprotein complex might enhance the methylation of the TPPP promoter. Further study is needed to identify the factors implicated in this transcriptional regulation.
To better understand this regulatory paradigm, the authors overexpressed linc00899 in HeLa cells and in the retinal pigment epithelial cell line RPE1. While ectopic linc00899 overexpression did not significantly affect TPPP mRNA levels in either cell line, CRISPR system-mediated endogenous linc00899 overexpression reduced TPPP mRNA levels in RPE1 cells. This interesting result suggests that the subnuclear site of linc00899 production affects function as a repressor of TPPP transcription.
Despite an escalation in knowledge of lncRNAs associated with cell proliferation, little is known about the lncRNAs directly implicated in controlling progression through the M-phase. In the comprehensive report by Stojic et al., the authors employed advanced and sophisticated approaches to identify prominent lncRNAs affecting precise segments of mitosis. By choosing informative readout parameters, the authors were able to identify, with high accuracy, the lncRNAs that regulated each step of mitosis. The authors further focused primarily on linc00899 and the underlying mechanism by which it delayed mitotic progression. In linc00899-depleted cells, TPPP was the most prominent differentially expressed gene. Although TPPP is a ubiquitous protein, it is most highly expressed in the brain, where linc00899 is remarkably low. Thus, the regulatory pathway described in the study by Stojic et al. might be particularly relevant to brain physiology and neurological diseases. In line with the suggested role for linc00899, a recent report uncovered that reducing linc00899 levels halted cell cycle entry in spinal ependymoma cells, albeit through the retinoblastoma-like protein 2 (RBL2)-dependent forkhead box protein (FoxO) pathway (22). Interestingly, linc00899 was recently reported to be highly abundant in the serum of acute myeloid leukemia (AML) patients (23); whether the linc00899-TPPP axis might govern mitosis impairment in AML warrants further investigation. While more complete catalogs of lncRNAs expressed in different cells and growth conditions are needed, the report by Stojic and colleagues charts a successful scheme for the identification of lncRNAs that govern specific stages of cellular division.
Acknowledgments
Funding: This work was funded in its entirety by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Xiya Lu, MD (Tongji Hospital of Tongji University, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri-2020-005). KA serves as an unpaid editorial board member of Non-coding RNA Investigation from April 2019 to March 2021. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484-8. [Crossref] [PubMed]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46. [Crossref] [PubMed]
- Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol 2016;937:3-17. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 2013;425:3723-30. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [Crossref] [PubMed]
- Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 2016;14:42-54. [Crossref] [PubMed]
- Kim J, Noh JH, Lee SK, et al. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget 2017;8:49409-20. [Crossref] [PubMed]
- Wang ZD, Shen LP, Chang C, et al. Long noncoding RNA lnc-RI is a new regulator of mitosis via targeting miRNA-210-3p to release PLK1 mRNA activity. Sci Rep 2016;6:25385. [Crossref] [PubMed]
- Marchese FP, Grossi E, Marin-Bejar O, et al. A long noncoding RNA regulates sister chromatid cohesion. Mol Cell 2016;63:397-407. [Crossref] [PubMed]
- Rošić S, Köhler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 2014;207:335-49. [Crossref] [PubMed]
- McIntosh JR. Mitosis. Cold Spring Harb Perspect Biol 2016;8:a023218 [Crossref] [PubMed]
- Juan G, Darzynkiewicz Z. Detection of mitotic cells. Curr Protoc Cytom 2004;Chapter 7:Unit 7 24.
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell 2004;7:45-60. [Crossref] [PubMed]
- Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol 2010;676:27-55. [Crossref] [PubMed]
- Stojic L, Lun ATL, Mascalchi P, et al. A high-content RNAi screen reveals multiple roles for long noncoding RNAs in cell division. Nat Commun 2020;11:1851. [Crossref] [PubMed]
- Kim S, Rhee K. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One 2014;9:e87016 [Crossref] [PubMed]
- Tirián L, Hlavanda E, Oláh J, et al. TPPP/p25 promotes tubulin assemblies and blocks mitotic spindle formation. Proc Natl Acad Sci U S A 2003;100:13976-81. [Crossref] [PubMed]
- Tokési N, Lehotzky A, Horváth I, et al. TPPP/p25 promotes tubulin acetylation by inhibiting histone deacetylase 6. J Biol Chem 2010;285:17896-906. [Crossref] [PubMed]
- Vitre B, Coquelle FM, Heichette C, et al. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol 2008;10:415-21. [Crossref] [PubMed]
- Barr AR, Gergely F. MCAK-independent functions of ch-Tog/XMAP215 in microtubule plus-end dynamics. Mol Cell Biol 2008;28:7199-211. [Crossref] [PubMed]
- Chen QB, Li ZH, Fu Y, et al. Downregulated long non-coding RNA LINC00899 inhibits invasion and migration of spinal ependymoma cells via RBL2-dependent FoxO pathway. Cell Cycle 2019;18:2566-79. [Crossref] [PubMed]
- Wang Y, Li Y, Song HQ, et al. Long non-coding RNA LINC00899 as a novel serum biomarker for diagnosis and prognosis prediction of acute myeloid leukemia. Eur Rev Med Pharmacol Sci 2018;22:7364-70. [PubMed]
Cite this article as: Tsitsipatis D, Amarbat B, Gorospe M, Abdelmohsen K. High-precision screen to identify lncRNAs governing specific mitotic stages. Non-coding RNA Investig 2020;4:9.


