
A microRNA prevents resistance to targeted therapy in chronic myeloid leukemia
The treatment of chronic phase chronic myeloid leukemia (CML) was revolutionized by the introduction of imatinib mesylate and later generations of tyrosine kinase inhibitors (TKIs) that potently inhibit the chimeric BCR-ABL kinase, whose aberrant activity both defines CML and underlies its molecular pathogenesis (1). These novel compounds helped establish a new paradigm for cancer treatment focused on targeted therapies that specifically inhibit the dysregulated or hyperactive signaling kinases that frequently drive malignant progression, a strategy which has now been successfully applied to many other hematologic malignancies and solid tumors. Nevertheless, many challenges still remain in treating CML patients including selection of best initial therapy based on patient- and disease-specific characteristics, managing acquired resistance to therapy, and more effectively targeting the leukemic stem cell population to achieve better disease control and potentially a cure. Currently, initial treatment is generally guided by validated risk scores such as the Sokal score or the Hasford score which predict the aggressiveness of disease, and are determined by the patient’s age and clinical features such as spleen size and various hematologic parameters such as blast count and platelet count (2,3). The first generation TKI, imatinib, is generally reserved for patients with low-risk disease, while the second generation TKIs, bosutinib, dasatinib, or nilotinib, are often preferred initial therapies for patients with intermediate- or high-risk disease, though other factors such as patient age and comorbidities and drug side effect profiles also inform treatment selection (4-7). Response to treatment is assessed by monitoring BCR-ABL transcript levels and, if these transcript levels either do not decline as expected or begin to increase, then evaluating for acquired mutations in BCR-ABL that are associated with resistance to particular TKIs, which would prompt a change in TKI therapy. This mutation profiling is epitomized by the T315I mutation in BCR-ABL which predicts resistance to all TKIs except the third generation TKI, ponatinib (7-9). Notably, molecular features such as gene expression or microRNA profiles have not been included in risk stratification, therapeutic resistance profiling, or treatment selection, though emerging research shows their great potential in both CML and other malignancies.
A rapidly expanding literature on the contributions of microRNAs to the development and progression of myeloid malignancies, including CML and acute myeloid leukemia (AML), continues to reveal new microRNA targets and their pathologic mechanisms, such as recent reports demonstrating the crucial function of miR-155 in FLT3-ITD AML (10,11). Multiple molecular events can cause microRNA dysregulation in myeloid malignancies, including dysregulated microRNA processing, gene deletion or mutation, or epigenetic alterations, all of which can have potential consequences for disease behavior and prognosis (12). Several studies investigating the roles of microRNAs in CML have focused on those that regulate BCR-ABL expression (13,14) or vice versa (15,16). Other studies have evaluated associations between microRNA expression and responsiveness to TKI therapy, including one recent publication that identified changes in miR-21 and miR-451 expression levels as predictive of TKI sensitivity in CML patients (17).
In the recent paper published in Blood by Lin and Rothe et al. (18), the authors perform deep sequencing of microRNAs to find not only differential expression of microRNAs between normal control bone marrow cells and leukemic cells from CML patients, but they also compare differential expression in CML patients who responded to TKI therapy (“responders”) and those who did not (“non-responders”) with the goal of identifying microRNAs as both predictive biomarkers of TKI sensitivity and to help elucidate potential microRNA-mediated mechanisms of TKI resistance for possible therapeutic exploitation. Here, and throughout most of their experiments involving patient samples, they utilized CD34+ cells, enriched for immature leukemic stem cells—rather than bulk leukemic cells—as their aim is to uncover mechanisms of TKI resistance in this particular population which is most often treatment-refractory (19). Using this strategy, they identify miR-185 as having lower expression levels in TKI-non-responders compared to TKI-responders, which they go on to confirm in additional cohorts of 22 retrospective CML patients and 58 prospective CML patients. To investigate the possible role of miR-185 in mediating TKI sensitivity, they demonstrate that CD34+ cells from TKI-non-responding patients have increased sensitivity to TKIs in in vitro colony forming assays when transduced with miR-185 compared to empty vector transduction. They further show that ectopic expression of miR-185 enhances TKI sensitivity in xenograft models using the human CML cell line BV173 and patient samples from TKI-non-responders, which resulted in decreased disease burden and improved survival of recipient mice. Using gene set enrichment analysis, they identified differences in oxidative phosphorylation and reactive oxygen species (ROS) pathways between non-responders and responders suggesting an association between metabolic perturbations and TKI resistance. With inversely correlated gene expression profiling they found PAK6 as a potential target gene of miR-185 that has increased expression in TKI-non-responders, and which appears to mediate TKI resistance through enhanced MAPK signaling. Finally, they show that combined inhibition of PAK6 with PF-3758309 and BCR-ABL with first and second generation TKIs results in reduced mitochondrial activity and ROS production as well as decreased leukemic cell viability and colony formation. Thus, they conclude that reduced levels of miR-185 can be used to predict which CML patients are likely to have substandard responses to TKI therapy and that this TKI resistance can be potentially overcome by simultaneous inhibition of PAK6, whose oncogenic activity is derepressed in the setting of low miR-185 levels (Figure 1).
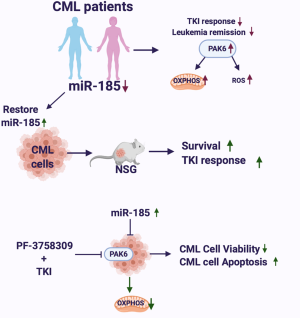
In this study the authors’ use of CML patient samples and their correlation of patient outcomes with novel molecular findings exemplify the goals of translational research and the “bedside to bench and back again” approach. By examining microRNA levels in the samples of more than 80 CML patients they were able to identify reduced miR-185 expression as a biomarker of inadequate response to TKI therapy and further to show in in vitro assays and in vivo xenograft models that restored expression of miR-185 can confer enhanced TKI sensitivity. Thus, their findings are not only predictive of treatment response but help elucidate a novel mechanism of TKI resistance, which involves miR-185-dependent regulation of the serine/threonine kinase PAK6 and dysregulated mitochondrial activity and ROS production, such that pharmacologic inhibition of PAK6 can cooperatively inhibit leukemic cell growth and survival with traditional TKIs in otherwise TKI-resistant cells. There are a number of details in the authors’ proposed model of a miR-185-PAK6-mitochondrial dysregulation axis that remained to be fully characterized. That CML cells—especially those exhibiting therapy resistance—might have enhanced or dysregulated mitochondrial oxidative phosphorylation and ROS production is not necessarily surprising, and there may be other contributing factors to these metabolic aberrancies besides enhanced PAK6 activity, which might also represent potential targetable mechanisms of TKI resistance. Further, it is unclear how PAK6 influences mitochondrial activity in this setting. The authors’ emphasis on leukemic stem cells and consistent use of CD34+ cells from patient samples represents another strength of this paper, as this immature population has long been associated with therapy resistance both in CML and other myeloid malignancies (19). However, the role of the tumor microenvironment and bone marrow niche in mediating therapy resistance in hematologic malignancies is becoming increasingly more appreciated (20-22), and in vitro assays and in vivo xenograft models designed to explore cell intrinsic mechanisms of TKI resistance such as differential microRNA expression can unfortunately ignore cell extrinsic phenomena such as microenvironment interactions. Patient samples can obviously provide a wealth of relevant information about coding and non-coding gene expression profiles but are understandably limited as models for evaluating microenvironment interactions. Finally, it would be interesting to know what mechanisms exist for determining the differential expression of miR-185 in different CML patients; somewhat counterintuitively, the authors show that inhibition of BCR-ABL with TKIs can increase miR-185 expression, suggesting that BCR-ABL activity could be perhaps one of several factors contributing to suppressed miR-185 levels that would be at least partly relieved by TKI therapy.
Chronic phase CML can at first appear to be a fairly homogenous disease defined by the translocation t(9;22) that produces the oncogenic fusion protein BCR-ABL kinase, however the clinically observed differences in TKI sensitivity strongly suggest the existence of significant molecular heterogeneity that determine the disease’s behavior and response to therapy in individual cases. As we learn increasingly more about the role of microRNAs in various hematologic malignancies and solid tumors, we are finding that microRNAs and other non-coding RNAs are crucial contributors to this molecular heterogeneity in cancers, and accordingly, they hold great value for prognosis and predicting responses to available therapies, as well as revealing potential targets for new treatments or overcoming resistance to old ones. For example, with regard to CML, one could envision in the near future the inclusion of microRNA profiling into risk assessment scores such as a modified Sokal score for the selection of best initial treatment at diagnosis, or being used to determine which patients might benefit from the addition of other inhibitors to standard TKI therapy. Many questions remain, however, such as the links between microRNAs and regulation of cellular metabolic processes, particularly in key populations such as cancer stem cells or cancer cells that acquire metastatic potential. Also unclear are the mechanisms by which key microRNAs become dysregulated and what role tumor microenvironment interactions might play in determining microRNA function. Additionally, while this paper introduces miR-185 as a mediator of inherent resistance to TKI therapy present at the time of diagnosis and persisting for at least three months after initiation of therapy, it will be important to explore microRNA profiles in patients who acquire therapy resistance and lose TKI sensitivity over time or who transition into the accelerated phase of disease or into blast crisis, particularly in those cases that do not harbor acquired resistance mutations in the BCR-ABL gene. A thorough study of the clonal evolution of resistant subpopulations could potentially yield important insights into the dynamics of microRNA expression and the selection pressures that define the regulation of their function in malignant progression. Finally, as more data accumulates on the role of microRNAs in the pathogenesis of myeloid malignancies, it will be interesting to see what similarities and differences exist in the microRNA landscapes of CML and AML which might have potential therapeutic implications for both diseases. Especially within the broad genetic and epigenetic heterogeneity of AML, it will be important to observe any emerging patterns of microRNA expression and function that may reveal common therapeutic resistance mechanisms or even potential new targets that may lead to novel treatment strategies for patients with myeloid malignancies.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Kai Wang, PHD (The First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri-2020-006). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Faderl S, Talpaz M, Estrov Z, et al. The biology of chronic myeloid leukemia. N Engl J Med 1999;341:164-72. [Crossref] [PubMed]
- Sokal JE, Cox EB, Baccarani M, et al. Prognostic discrimination in "good-risk" chronic granulocytic leukemia. Blood 1984;63:789-99. [Crossref] [PubMed]
- Hasford J, Pfirrmann M, Hehlmann R, et al. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa. Writing Committee for the Collaborative CML Prognostic Factors Project Group. J Natl Cancer Inst 1998;90:850-8. [Crossref] [PubMed]
- Cortes JE, Saglio G, Kantarjian HM, et al. Final 5-Year Study Results of DASISION: The Dasatinib Versus Imatinib Study in Treatment-Naive Chronic Myeloid Leukemia Patients Trial. J Clin Oncol 2016;34:2333-40. [Crossref] [PubMed]
- Cortes JE, Gambacorti-Passerini C, Deininger MW, et al. Bosutinib Versus Imatinib for Newly Diagnosed Chronic Myeloid Leukemia: Results From the Randomized BFORE Trial. J Clin Oncol 2018;36:231-7. [Crossref] [PubMed]
- Larson RA. Is there a best TKI for chronic phase CML? Hematology Am Soc Hematol Educ Program 2015;2015:250-6. [Crossref] [PubMed]
- Radich JP, Deininger M, Abboud CN, et al. Chronic Myeloid Leukemia, Version 1.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2018;16:1108-35. [Crossref] [PubMed]
- Patel AB, O'Hare T, Deininger MW. Mechanisms of Resistance to ABL Kinase Inhibition in Chronic Myeloid Leukemia and the Development of Next Generation ABL Kinase Inhibitors. Hematol Oncol Clin North Am 2017;31:589-612. [Crossref] [PubMed]
- Shanmuganathan N, Hughes TP. Molecular monitoring in CML: how deep? How often? How should it influence therapy? Hematology Am Soc Hematol Educ Program 2018;2018:168-76. [Crossref] [PubMed]
- Wallace JA, Kagele DA, Eiring AM, et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood 2017;129:3074-86. [Crossref] [PubMed]
- O'Connell RM, Rao DS, Chaudhuri AA, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med 2008;205:585-94. [Crossref] [PubMed]
- Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood 2017;130:1290-301. [Crossref] [PubMed]
- Xishan Z, Ziying L, Jing D, et al. MicroRNA-320a acts as a tumor suppressor by targeting BCR/ABL oncogene in chronic myeloid leukemia. Sci Rep 2015;5:12460. [Crossref] [PubMed]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 2008;13:496-506. [Crossref] [PubMed]
- Zhang B, Nguyen LXT, Li L, et al. Bone marrow niche trafficking of miR-126 controls the self-renewal of leukemia stem cells in chronic myelogenous leukemia. Nat Med 2018;24:450-62. [Crossref] [PubMed]
- Srutova K, Curik N, Burda P, et al. BCR-ABL1 mediated miR-150 downregulation through MYC contributed to myeloid differentiation block and drug resistance in chronic myeloid leukemia. Haematologica 2018;103:2016-25. [Crossref] [PubMed]
- Alves R, Goncalves AC, Jorge J, et al. MicroRNA signature refine response prediction in CML. Sci Rep 2019;9:9666. [Crossref] [PubMed]
- Lin H, Rothe K, Chen M, et al. The miR-185/PAK6 Axis Predicts Therapy Response and Regulates Survival of Drug-Resistant Leukemic Stem Cells in CML. Blood 2020;136:596-609. [Crossref] [PubMed]
- Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood 2017;129:1595-606. [Crossref] [PubMed]
- Medyouf H. The microenvironment in human myeloid malignancies: emerging concepts and therapeutic implications. Blood 2017;129:1617-26. [Crossref] [PubMed]
- Qu Y, Dou B, Tan H, et al. Tumor microenvironment-driven non-cell-autonomous resistance to antineoplastic treatment. Mol Cancer 2019;18:69. [Crossref] [PubMed]
- Tabe Y, Konopleva M. Leukemia Stem Cells Microenvironment. Adv Exp Med Biol 2017;1041:19-32. [Crossref] [PubMed]
Cite this article as: Ghazaryan A, Goodwin CB, O’Connell RM. A microRNA prevents resistance to targeted therapy in chronic myeloid leukemia. Non-coding RNA Investig 2020;4:10.


