
Functional relevance of pyknons in tumor formation
In a recent Genome Biology paper, Rigoutsos et al. describe the functional characterization of a novel, pyknon-containing RNA. Their highly cooperative and pioneering work in pyknon and long non-coding RNA (lncRNA) research (1) indicates the role of the novel pyk-reg-90-containing lncRNA, novel pyk-reg-90-containing lncRNA (N-BLR), in cellular and molecular physiology as well as in the proliferation and metastasis-formation capabilities of colorectal cancer (CRC) cells. Well-grounded in silico, in vitro and in situ results are supported by patient survival data.
Although the majority of the human genome is transcribed into RNA, only approximately 2% of these transcripts are translated into proteins. RNA molecules that lack protein-coding potential are collectively referred to as non-coding (nc) RNAs. Various experimental approaches have identified several classes of ncRNAs, including microRNAs, Piwi-interacting RNAs, sno-derived RNAs, transfer RNA fragments, lncRNAs and others. LncRNAs have been drawing increasing attention in recent years as they have been found to be differently expressed in various diseased conditions. However, whether all these long non-coding transcripts are functional or just transcriptional noise is still a controversial unknown (2): functional analysis has been performed only on less than 1% of all described lncRNAs.
By introducing a novel method for computational identification of DNA sequence motifs a decade ago, Rigoutsos et al. identified short (≥16 nucleotides) DNA sequence motifs called pyknons, which are enriched in both protein-coding and untranslated regions of genes (≥30 intact copies) (3,4). Although interspecies conservation of pyknon sequences has not been detected, copies are enriched in groups of protein-coding genes involved in the same biological processes in two species, suggesting that their functions might be conserved (4).
Repetitive DNA elements are estimated to comprise 66–69% of the human genome (5) and are thought to have a role in evolution (6). Despite this abundant presence in the genome, we still have little knowledge about their physiological relevance and molecular regulation (7), although some repeat sequences have been shown to function as gene-regulatory elements and promoters (8).
The vast majority of pyknons were found to overlap with classical repeat elements; although approximately 50,000 pyknons are found in repeat-free regions. Pyknons are generally smaller than retroviral elements (e.g., Alu elements), and there is a possibility that they originate from transposable elements (3).
With the aim of elucidating the functional role(s) of pyknons, which form one sixth of the human genome, Rigoutsos et al. (1) describe the functional analysis of pyknons in CRC cells (3). After an initial expression analysis of several pyknons in CRC cell lines and CRC samples, they selected one pyknon (pyk-reg-90) to study in more depth. The high expression level of this pyknon was associated with poor overall survival in CRC patients in independent cohorts.
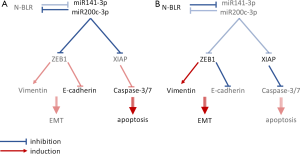
They found a pyk-reg-90 instance in Chr. 3p21 to be located in a novel lncRNA, N-BLR. Initial characterization of N-BLR revealed higher expression levels in both primary colorectal tumor cells and metastases and localization in both the nucleus and cytoplasm. To functionally analyze N-BLR in cellular functions, N-BLR knock-down experiments were carried out and resulted in a reduction in cell number and upregulation of apoptosis. These events were accompanied by an upregulation of apoptotic markers (caspase-3/7, caspase-8 and caspase-9 and cleaved PARP-1), whereas X-liked inhibitor of apoptosis (XIAP) was downregulated.
Since N-BLR is also highly expressed in metastasizing tumor cells, its contribution to migration and invasion was studied. N-BLR knock-down cells showed decreased invasion and migratory capability with a decrease in vimentin levels and upregulated E-cadherin. Moreover, opposite effects were seen in cells overexpressing N-BLR. A transcription factor known to facilitate cell migration and invasion, zinc finger E-box binding homeobox 1 (ZEB-1), is involved in the epidermal-mesenchymal transition (EMT) of tumor cells. ZEB-1 is a negative regulator of E-cadherin expression and a positive regulator of vimentin expression and is regulated by the miR-200 family through a negative feedback loop (9). Since ZEB-1 expression decreased in N-BLR knock down cells, Rigoutsos et al. supposed an interaction between N-BLR and members of the miR-200 family. They predicted and confirmed N-BLR interaction with two miR-200 family members: miR-141-3p and miR-200c-3p. Expression levels of these two miRNAs increased in N-BLR knock down cells and decreased in N-BLR overexpressing cells, whereas transfection with miR-141-3p and miR-200c-3p mimics decreased N-BLR levels. Similarly, when analyzing expression levels in tumor samples, an inverse correlation between levels of miR-141-3p/miR-200c-3p and N-BLR was found, suggesting a negative feedback loop between the miRNAs and the lncRNA (Figure 1). In an analysis of the interaction sequences of N-BLR and miR-200c-3p, the 20 nt pyknon motif was found to be part of the interaction site. The importance of the miR-200c-3p–N-BLR axis in tumor cell formation was moreover demonstrated by the ability of these molecules to regulate 5-fluoro-uracil sensitivity/resistance in CRC cells.
Although pyknon-like DNA motifs have been described in several genomes (3,4,10), their roles have not been researched as well as for the human genome. In this set of impressive experiments, Rigoutsos et al. demonstrated that N-BLR is a marker for poor disease prognosis in colorectal carcinoma as well as a regulator of tumor progression and EMT through a direct interaction with miR-141-3p and miR-200c-3p. Their results provided the first evidence for the functionality of pyknons and pyknon-containing sequences. To determine whether other pyknons are also functional, the pyknon database (http://cm.jefferson.edu/pyknons.html) could be used to analyze recently described sequence-specific lncRNA interactions with mRNAs (11,12), miRNAs (13-16) or genomic DNA sequences (17) to determine whether the lncRNAs include pyknon sequences.
Perhaps the most significant aspect of the Rigoutsos et al. paper is that it opens the possibility that pyknons could be used to locate functional lncRNAs, as was the case for N-BLR. Such a method might address one of the biggest challenges in lncRNA biology: to distinguish between functional and “junk” transcripts (2).
Acknowledgments
Funding: This work was supported by the National Research Development and Innovation Office (GINOP-2.3.2-15-2016-00015).
Judit Danis was supported by the NTP-NFTÖ-17 project provided by the Hungarian Ministry of Human Capacities.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Section Editor Jinzhe Zhou (Department of General Surgery, Tongji Hospital, Tongji University School of Medicine, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2017.12.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rigoutsos I, Lee SK, Nam SY, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol 2017;18:98. [Crossref] [PubMed]
- Palazzo AF, Lee ES. Non-coding RNA: What is functional and what is junk? Front Genet 2015;6:2. [PubMed]
- Rigoutsos I, Huynh T, Miranda K, et al. Short blocks from the noncoding parts of the human genome have instances within nearly all known genes and relate to biological processes. Proc Natl Acad Sci U S A 2006;103:6605-10. [Crossref] [PubMed]
- Tsirigos A, Rigoutsos I. Human and mouse introns are linked to the same processes and functions through each genome’s most frequent non-conserved motifs. Nucleic Acids Res 2008;36:3484-93. [Crossref] [PubMed]
- de Koning AP, Gu W, Castoe TA, et al. Repetitive Elements May Comprise Over Two-Thirds of the Human Genome. PLoS Genet 2011;7:e1002384 [Crossref] [PubMed]
- Jurka J. Evolutionary impact of human Alu repetitive elements. Curr Opin Genet Dev 2004;14:603-8. [Crossref] [PubMed]
- Padeken J, Zeller P, Gasser SM. Repeat DNA in genome organization and stability. Curr Opin Genet Dev 2015;31:12-9. [Crossref] [PubMed]
- Göke J, Ng HH. CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep 2016;17:1131-44. [Crossref] [PubMed]
- Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 2014;16:488-94. [Crossref] [PubMed]
- Feng J, Naiman DQ, Cooper B. Coding DNA repeated throughout intergenic regions of the Arabidopsis thaliana genome: evolutionary footprints of RNA silencing. Mol Biosyst 2009;5:1679-87. [Crossref] [PubMed]
- Wu X, Yan T, Wang Z, Wu X, Cao G, Zhang C. LncRNA ZEB2-AS1 promotes bladder cancer cell proliferation and inhibits apoptosis by regulating miR-27b. Biomed Pharmacother 2017;96:299-304. [Crossref] [PubMed]
- Danis J, Göblös A, Bata-Csörgő Z, et al. PRINS Non-Coding RNA Regulates Nucleic Acid-Induced Innate Immune Responses of Human Keratinocytes. Front Immunol 2017;8:1053. [Crossref] [PubMed]
- Ren Y, Shang J, Li J, et al. The long noncoding RNA PCAT-1 links the microRNA miR-215 to oncogene CRKL-mediated signaling in hepatocellular carcinoma. J Biol Chem 2017;292:17939-49. [Crossref] [PubMed]
- Liu W, Ma C, Yang B, et al. LncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat deposition and contribute to body weight gain in mice. Biochem Biophys Res Commun 2017;493:1168-75. [Crossref] [PubMed]
- Zhao L, Sun H, Kong H, et al. The Lncrna-TUG1/EZH2 Axis Promotes Pancreatic Cancer Cell Proliferation, Migration and EMT Phenotype Formation Through Sponging Mir-382. Cell Physiol Biochem 2017;42:2145-58. [Crossref] [PubMed]
- Yan X, Zhang D, Wu W, et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and miR-34a. Cancer Res 2017;77:6704-16. [Crossref] [PubMed]
- Jalali S, Singh A, Maiti S, et al. Genome-wide computational analysis of potential long noncoding RNA mediated DNA:DNA:RNA triplexes in the human genome. J Transl Med 2017;15:186. [Crossref] [PubMed]
Cite this article as: Danis J, Széll M. Functional relevance of pyknons in tumor formation. Non-coding RNA Investig 2018;2:3.


