
TWIST1 regulation of circRNA: a novel mechanism to promote epithelial-mesenchymal transition in hepatocellular carcinoma
The epithelial-mesenchymal transition (EMT) is a process in which epithelial cells reversibly transdifferentiate and adopt a mesenchymal morphology (1-3). EMT is driven by suppression of epithelial associated genes such as E-cadherin and upregulation of mesenchymal associated genes such as fibroblast-specific protein 1 and Vimentin (1-3). These genetic changes lead epithelial cells to become more spindle-like in appearance due to loss of epithelial cell apical-basal polarity, cell-cell interactions, and cell-extracellular matrix interactions (1). In addition to a mesenchymal morphology, EMT confers cells with increased motility and invasiveness (1-3). EMT is an essential process for development, specifically gastrulation and neural crest migration, wound healing, and tissue regeneration (1). This developmental process is hijacked by tumor cells to mediate invasion-metastasis, cancer cell stemness, and therapeutic resistance (1-3). EMT-transcription factors (EMT-TFs), such as TWIST1, are the main mediators of EMT as they induce transcriptional programs that leads to repression of numerous epithelial-associated genes and induction of mesenchymal-associated genes as well as regulating genes that promote survival and proliferation (1). For example, the EMT-TF, ZEB1 directly represses E-cadherin expression and induces expression of N-cadherin and Vimentin (1,4). Similarly, TWIST1 can directly suppress E-cadherin expression by binding to E-box binding sites within the CDH1 (E-cadherin gene) promoter and recruiting repression complexes (5). TWIST1 can also indirectly suppress E-cadherin expression by upregulating other EMT-TFs such as SNAI2 which then directly binds the CDH1 promoter and represses CDH1 transcription (6). Given the pleotropic transcriptional program induced by EMT-TFs, EMT-TF expression has been implicated in tumorigenesis due to their ability to induce EMT, suppress apoptosis, suppress oncogene-induced senescence (OIS), and induce therapeutic resistance (7). It is currently unclear if the ability of EMT-TFs to inhibit failsafe programs like apoptosis and mediate therapeutic resistance is dependent on their ability to induce EMT. Additionally, while EMT-TFs have been shown to directly regulate EMT-related genes like CDH1 and VIM directly, it remains unclear if EMT-TFs regulate the EMT program through other indirect mechanisms.
Circular RNA (circRNA) is a recently identified form of non-coding RNA that results from alternative splicing of pre-mRNA (8,9). In order to form circRNA, a 3' or downstream splice donor site is “back spliced” to a 5' splice site resulting in a 3'-5' covalently linked circularized form of RNA (8,9). Interestingly, expression of numerous circRNAs has been implicated in tumorigenesis and have been shown to promote tumor proliferation, growth, invasion and migration in a variety of solid tumor types (9). Additionally, expression of circRNA is typically associated with later stage tumors and poor patient prognosis (9). circRNA can promote tumorigenesis through multiple mechanisms. circRNA often have multiple miRNA binding sites and the main mechanism by which circRNA promote tumorigenesis is by functioning as a miRNA “sponge” (9). For example, in lung cancer, expression of Hsa_circ_0012673 promotes growth of lung cancer cells by binding a miRNA that inhibits Erb-b2 receptor tyrosine kinase 3 expression (10,11). Alternatively, circRNA can directly bind proteins and subsequently promote protein-protein interaction, alter localization of proteins, and alter transcription of numerous genes (11). For example, circ-Amotl1 can directly bind to c-Myc and induce its nuclear localization and promote expression of c-Myc target genes (9,11). Additionally, circRNA have been found to stimulate transcription of the genes from which they are derived by binding to RNA polymerase II and through poorly characterized mechanisms induce transcription (12). Interestingly, some circRNA have the necessary elements needed for translation, however, it is unclear if circRNA encoded peptides play a significant role in circRNA-mediated tumorigenesis (9).
Recent studies have suggested that one of the mechanisms by which circRNA expression may promote tumorigenesis is by inducing EMT. For example, in bladder carcinoma cells, expression of circRNA-MYLK induces EMT by inhibiting mir-29a and subsequently activating VEGFA/VEGFR2 (13). Additionally, expression of hsa_circ_0007534 in non-small cell lung cancer (NSCLC) has oncogenic properties in part by inducing EMT (14). Despite recent studies demonstrating circRNA expression leads to EMT, the mechanisms by which circRNA promotes EMT remain largely unknown.
Interestingly, a recent report by Meng et al. provided the first evidence demonstrating a role for circRNA in mediating EMT and tumorigenesis promoted by EMT-TFs. In this study, the authors demonstrated that in hepatocellular carcinoma (HCC) cells, TWIST1 increases Cullin2 (CUL2) promoter activity by directly binding to the CUL2 promoter (15). TWIST1 binding at the CUL2 promoter actually resulted in downregulation of CUL2 mRNA and protein, while specifically inducing expression of the CUL2 circRNA, circ-10720 (Figure 1) (15). The authors demonstrate that expression of circ-10720 phenocopies TWIST1 expression in HCC cells by downregulating E-cadherin, upregulating of Vimentin, and increasing migration and invasion (15). The authors identify a mechanism through which TWIST1 induces Vimentin by demonstrating that circ-10720 can lead to Vimentin upregulation by binding and inhibiting miRs (miRs-1246, -578, and -490-5p) that inhibit Vimentin expression (Figure 1) (15). Additionally, in mouse models of TWIST1-mediated HCC, the authors found that silencing circ-10720 expression inhibited TWIST1-mediated metastasis, tumor growth, and Vimentin expression (15). Overall, Meng et al. provide clear evidence that one of the key mechanisms by which the EMT-TF TWIST1 promotes EMT and HCC tumorigenesis is through regulating circRNA. Additionally, the authors provide the first mechanistic insight into how TWIST1 expression leads to upregulation of Vimentin.
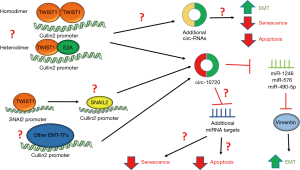
Meng et al. identified a novel mechanism by which TWIST1 can indirectly regulate gene expression, via direct upregulation of circRNA. While the authors demonstrated that circ-10720 is a critical target for TWIST1-mediated HCC tumorigenesis, it remains unclear if TWIST1 regulates other circRNAs in HCC and if these potential targets also play a role in TWIST1-mediated tumorigenesis (Figure 1). As EMT-TFs often display tissue specific phenotypes (16), future investigations should be aimed at determining if this phenotype is relevant for TWIST1-mediated tumorigenesis in other solid tumors. Furthermore, it is unknown whether TWIST1 regulation of circRNA is a TWIST1-specific phenomenon or will be observed with other EMT-TFs. Given that EMT-TFs share a response element (E-box binding site), numerous target genes, and often have similar functions, it is likely that circ-10720 and other circRNAs are regulated by other EMT-TFs (Figure 1). Of note, previous studies have demonstrated that EMT-TFs can regulate Vimentin expression, for example, ZEB1 has been previously shown to induce Vimentin expression by binding the chromatin remodeling protein BRG1 and likely directly upregulating Vimentin transcription (4). However, EMT-TFs often have multiple mechanisms of regulating a single gene, thus it is still plausible that ZEB1 upregulation of circ-10720 may play a role in ZEB1 upregulation of Vimentin. TWIST1 has also been shown to indirectly regulate genes involved in EMT, such as E-cadherin, by upregulating other EMT-TFs, specifically SNAI2 (6). Future studies should be aimed at determining if TWIST1 upregulation of other EMT-TFs, specifically SNAI2, contributes to TWIST1 upregulation of circ-10720 (Figure 1).
Previous studies have demonstrated that TWIST1 has the ability form TWIST1-TWIST1 homodimers or heterodimers with the E2A proteins (17). The TWIST1 homodimer and heterodimer often regulate distinct sets of TWIST1 target genes and have been shown to have divergent, often antagonist, functions (17). Our group recently demonstrated that known target genes of TWIST1, YBX1 and SNAI2, were only induced with the TWIST1-E2A heterodimer (18). Additionally, the TWIST1-E2A heterodimer seems to be critical for TWIST1-mediated NSCLC tumorigenesis (18). Others have demonstrated in breast and prostate cancer that the TWIST1-E2A heterodimer is required for TWIST1 suppression of OIS and TWIST1-mediated metastasis (19,20). Conversely, other studies have demonstrated that the TWIST1 homodimer is critical for TWIST1-mediated EMT and chemoresistance in pancreatic cancer (21). Meng et al. demonstrate that TWIST1 selectively induces circ-10720 while repressing CUL2 mRNA expression in HCC cells. Future studies should be aimed at determining if this TWIST1 function is dependent on a specific TWIST1 binding partner (Figure 1) and how binding at the upstream promoter leads to specific induction of circ-10720. Of note, a recent study demonstrated that the small molecule, harmine, can inhibit TWIST1 function both in vitro and in vivo by selectively degrading the TWIST1-E2A binding partner (18). If TWIST1 upregulation of circ-10720 requires the TWIST1-E2A heterodimer, harmine may be a potential therapeutic option to treat TWIST1-driven HCC. Targeting TWIST1 may prove to be a more viable therapeutic option than targeting circ-10720 as efficacious studies using therapies targeting circRNA are currently lacking (9).
An important aspect of EMT-TFs is their ability to promote tumorigenesis through EMT-independent mechanisms (7). For example, in both breast cancer and NSCLC, TWIST1 is required for tumorigenesis by inhibiting OIS and/or apoptosis (18,20,22-24). While Meng et al. clearly demonstrate that circ-10720 expression induces Vimentin expression and subsequently EMT, it remains unclear if additional targets of circ-10720 play a role in promoting these EMT-independent functions of TWIST1 and/or other EMT-TFs (Figure 1). Given that Meng et al. demonstrate that inhibition of circ-10720 inhibits both metastasis and tumor proliferation, it is likely that Vimentin is not the only important target of circ-10720. Of note, EMT-TFs, such as ZEB1 and TWIST1 have been shown to suppress apoptosis and mediate therapeutic resistance by suppressing transcription of the pro-apoptotic protein, BIM (24,25). Future studies should be aimed at determining if circ-10720 negatively regulates pro-apoptotic genes or upregulates anti-apoptotic proteins. circ-10720 suppression of apoptosis is potentially targetable with recently developed small molecule inhibitors of BCL-2, BCL-2/BCL-xL, and MCL-1.
In summary, Meng et al. provide the first evidence that an EMT-TF can indirectly regulate genes by upregulating circRNA. Additionally, they provide the first mechanistic insight into how circRNA promotes EMT and how TWIST1 upregulates Vimentin expression. They also demonstrate that circ-10720 expression can partially phenocopy TWIST1 expression in HCC and is required for both tumorigenesis and metastasis in TWIST1-mediated models of HCC. This study suggests that both TWIST1 and/or circ-10720 may be viable targets in HCC and lay the groundwork for a new area of study: EMT-TF regulation of circRNAs.
Acknowledgments
Funding: ZA Yochum received support from the Howard Hughes Medical Institute Medical Fellowship, T32GM008421-22 and a 1F30CA213765-01.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Dr. Jin Li, Section Editor (Cardiac Regeneration and Ageing Lab, School of Life Sciences, Shanghai University, Shanghai, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ncri.2018.12.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. [Crossref] [PubMed]
- Nieto MA. Epithelial-Mesenchymal Transitions in development and disease: old views and new perspectives. Int J Dev Biol 2009;53:1541-7. [Crossref] [PubMed]
- Sanchez-Tillo E, Lazaro A, Torrent R, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010;29:3490-500. [Crossref] [PubMed]
- Vesuna F, van Diest P, Chen JH, et al. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun 2008;367:235-41. [Crossref] [PubMed]
- Casas E, Kim J, Bendesky A, et al. Snail2 is an Essential Mediator of Twist1-Induced Epithelial Mesenchymal Transition and Metastasis. Cancer Res 2011;71:245-54. [Crossref] [PubMed]
- Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 2014;16:488. [Crossref] [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [Crossref] [PubMed]
- Cui X, Wang J, Guo Z, et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer 2018;17:123. [Crossref] [PubMed]
- Wang X, Zhu X, Zhang H, et al. Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem Biophys Res Commun 2018;496:1069-75. [Crossref] [PubMed]
- Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ 2017;24:1609-20. [Crossref] [PubMed]
- Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as Therapeutic Agents and Targets. Front Physiol 2018;9:1262. [Crossref] [PubMed]
- Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 2017;403:305-17. [Crossref] [PubMed]
- Qi Y, Zhang B, Wang J, et al. Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo. Gene 2018;676:79-85. [Crossref] [PubMed]
- Meng J, Chen S, Han JX, et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res 2018;78:4150-62. [Crossref] [PubMed]
- Nieto MA. Context-specific roles of EMT programmes in cancer cell dissemination. Nat Cell Biol 2017;19:416-8. [Crossref] [PubMed]
- Ansieau S, Morel AP, Hinkal G, et al. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene 2010;29:3173. [Crossref] [PubMed]
- Yochum ZA, Cades J, Mazzacurati L, et al. A First-in-Class TWIST1 Inhibitor with Activity in Oncogene-Driven Lung Cancer. Mol Cancer Res 2017;15:1764-76. [Crossref] [PubMed]
- Gajula RP, Chettiar ST, Williams RD, et al. Structure-function studies of the bHLH phosphorylation domain of TWIST1 in prostate cancer cells. Neoplasia 2015;17:16-31. [Crossref] [PubMed]
- Jacqueroud L, Bouard C, Richard G, et al. The Heterodimeric TWIST1-E12 Complex Drives the Oncogenic Potential of TWIST1 in Human Mammary Epithelial Cells. Neoplasia 2016;18:317-27. [Crossref] [PubMed]
- Wang J, Nikhil K, Viccaro K, et al. The Aurora-A-Twist1 axis promotes highly aggressive phenotypes in pancreatic carcinoma. J Cell Sci 2017;130:1078-93. [PubMed]
- Burns TF, Dobromilskaya I, Murphy SC, et al. Inhibition of TWIST1 leads to activation of oncogene-induced senescence in oncogene-driven non-small cell lung cancer. Mol Cancer Res 2013;11:329-38. [Crossref] [PubMed]
- Tran PT, Shroff EH, Burns TF, et al. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet 2012;8:e1002650 [Crossref] [PubMed]
- Yochum ZA, Cades J, Wang H, et al. Targeting the EMT transcription factor TWIST1 overcomes resistance to EGFR inhibitors in EGFR-mutant non-small-cell lung cancer. Oncogene 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Song KA, Niederst MJ, Lochmann TL, et al. Epithelial-to-Mesenchymal Transition Antagonizes Response to Targeted Therapies in Lung Cancer by Suppressing BIM. Clin Cancer Res 2018;24:197-208. [Crossref] [PubMed]
Cite this article as: Yochum ZA, Burns TF. TWIST1 regulation of circRNA: a novel mechanism to promote epithelial-mesenchymal transition in hepatocellular carcinoma. Non-coding RNA Investig 2018;2:71.


